Fig. 4. The PfPCRCR complex bridges the parasite and erythrocyte membranes.
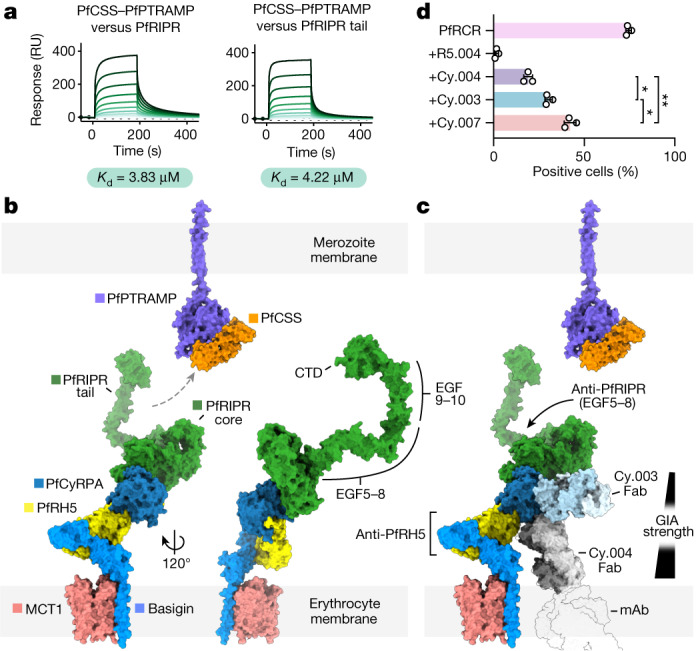
a, The PfCSS–PfPTRAMP complex binds to full-length PfRIPR and its tail with equal affinity, as measured by surface plasmon resonance; n = 4 independent experiments (one shown). RU, response units. The binding affinity given is the mean of those determined by steady-state analysis of each independent measurement (Extended Data Fig. 8d). b, Composite model of the PfRCR complex on the erythrocyte membrane, illustrating how the tail of PfRIPR projects towards the merozoite membrane where PfCSS–PfPTRAMP is located. EGF-like domains 5–8 and 9–10 and the CTD of PfRIPR are highlighted. The composite PfRCR complex model is aligned onto the structure of basigin-bound MCT1 (PDB: 7CKR33). For illustrative purposes only, the AlphaFold2-predicted structure of PfPTRAMP (magenta, AlphaFoldDB: Q8I5M8, residues 26–352) and the crystal structure of PfCSS (orange, PDB: 7UNY1) are docked together. c, Mapping of growth-inhibitory antibodies targeting PfRCR components in the context of erythrocyte binding. The potency of growth-inhibitory PfCyRPA-targeting antibodies correlates with their proximity to the erythrocyte membrane, illustrated by overlay of Cy.004 Fab-bound PfCyRPA (PDB: 7PHW5) onto PfRCR–Cy.003. The approximate location of the Fc portion of the intact monoclonal antibody is shown as a faint silhouette through overlay of the crystal structure of an intact antibody (PDB: 1IGT). Growth-inhibitory PfRIPR-targeting antibodies bind to EGF-like domains 5–8, located distal to the erythrocyte membrane. d, The ability of PfRCR to bind to human erythrocytes, either alone or in the presence of twofold molar excess of antibodies R5.004, Cy.003, Cy.004 and Cy.007, expressed as a percentage of PfRCR-positive cells; n = 3 independent measurements; mean plus or minus s.d. is shown; P = 0.0132 for Cy.003 versus Cy.004, P = 0.0418 for Cy.003 versus Cy.007 and P = 0.0023 for Cy.004 versus Cy.007 (one-way Brown–Forsythe and Welch analysis of variance adjusted for multiple comparisons with Dunnett T3).
