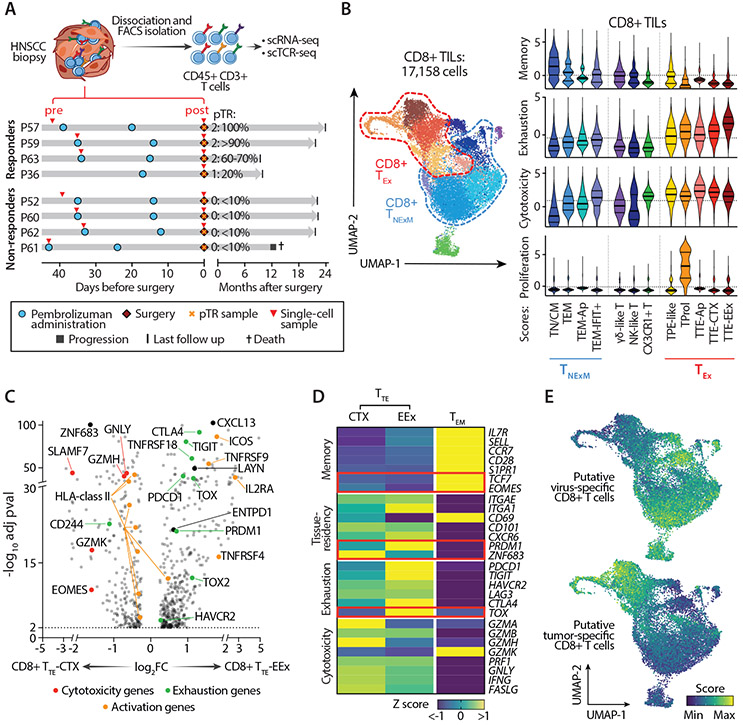
Fig. 2. Characterization of CD8+ TILs in HNSCC.
(A) Scheme of collection and processing of TILs for single-cell analysis. Clinical courses of 4 Rs and 4 NRs are represented. Red arrows - time of collection of the tumor biopsies profiled at baseline (pre) or at surgery after PD-1 blockade (post). pTR assessed at tumor resection is reported. (B) Classification of CD8+ HNSCC-TILs. UMAP of the transcriptionally-defined clusters from the scRNA-seq data (left), as denoted by colors. Right plots - The inferred cell states (x axis), colored to match the UMAP clusters. Cells from each cell state are scored for expression of memory, exhaustion, cytotoxicity and proliferation genes (see Materials and Methods), with medians and quartiles indicated (thick and thin horizontal lines, respectively). Horizontal dashed lines - average scores within the entire dataset of CD8+ TILs. (C) Differentially expressed genes for all cells in CD8+ TTE with cytotoxic characteristics (TTE-CTX) (left) and CD8+ TTE with extreme exhaustion programming (TTE-EEx) (right) clusters. Dots denote genes with adj-pval<0.01 (two-sided Wilcoxon Rank Sum test). Representative genes are labeled, with colors corresponding to gene-functions (legend). (D) Heatmap reporting variation (expressed as Z-score) in average gene-expression measured in exhausted TTE-CTX, TTE-EEx or control TEM CD8+ TILs. Qualitative differences are reported for representative T cell-related genes, divided based on their known function (left). Key transcriptional factors are highlighted in red. (E) UMAPs of CD8+ TILs colored based on enrichment of published gene-signatures associated with CD8+TILs with validated antiviral (top) or antitumor (bottom) reactivity (14). Abbreviations: TN/CM: naïve/central memory TILs; TEM: effector memory TILs; TPE-like: enriched in progenitor exhausted TILs; TProl: proliferative TILs; TTE: terminally exhausted TILs; CTX: cytotoxic; EEx: extremely exhausted; Ap: apoptotic; TNExM: non-exhausted memory TILs; TEx: Exhausted TILs.