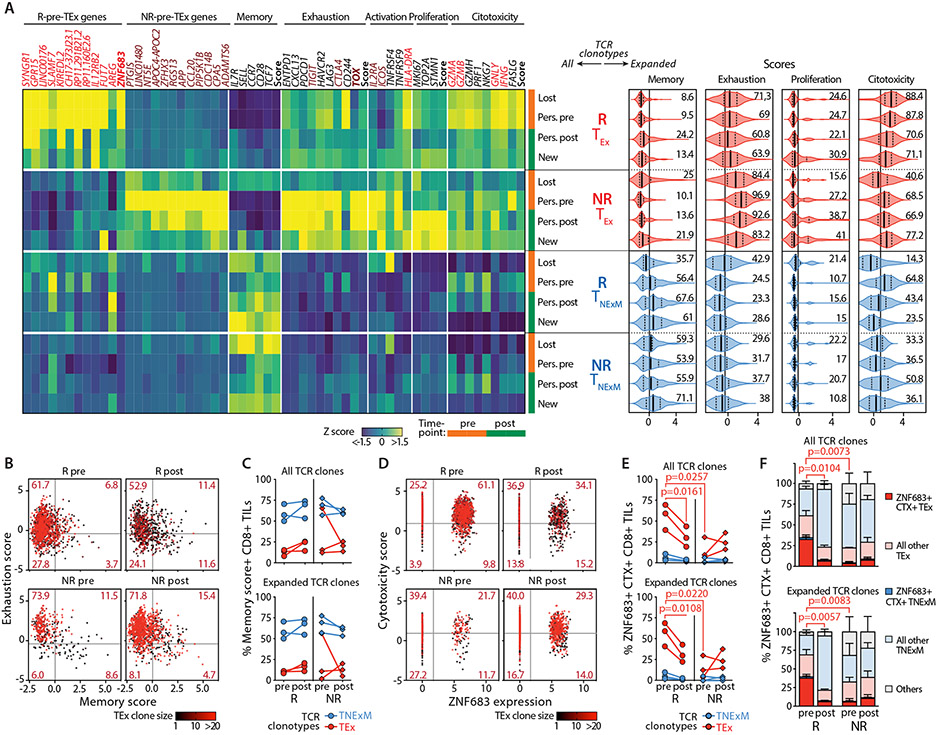
Fig. 7. High frequencies of ZNF683+ cytotoxic CD8+ TEx-TCR clonotypes associates with response to PD-1 blockade.
(A) Phenotypes of clones Lost/New/Persisting TCR clonotypes. A-Left: Gene-expression profile of TCR clonotypes (rows) classified based on their primary phenotype (TEx, TNExM) and divided based on their detection in pre and/or post-immunotherapy HNSCC tumors (Lost, New, Persisting [Pers.]) in Responders (R, n=3) or Non-Responders (NR, n=3). Average RNA-transcripts levels expressed as Z-scores are shown for i) top deregulated in R/NR-Pre-TEx signatures (see Data File S5); ii) for a panel of genes representative for T-cell related features; iii) for scores summarizing the expression of key genes (see Material and Methods). Genes enriched in R-Pre-TEx or NR-Pre-TEx signatures are labeled in red and brown respectively. Key transcriptional factors are highlighted in bold. Vertical tracks indicated evaluation of TCR clonotype in pre (orange) or post (green)-therapy timepoints. A-Right: Summary of the cell states of expanded (>2 cells) TCR clonotypes, divided by category (left). For each T-cell feature, scores summarize the expression of key genes (see Materials and Methods). Violin plots: median values (solid lines) with quartiles (dashed lines). Numbers: percentages of positive cells. Vertical lines: mean scores in the entire dataset. (B,C) Analysis of clonal revival in putative tumor reactive TEX-TCR clonotypes. (B) Bidimensional plot depicting quantification of memory and exhaustion programs (scores) in TEx-TCR clonotypes detected in HNSCC biopsies collected before (pre) or after (post) immunotherapy from 3Rs and 3 NRs. Dots represent single cells harboring TCRs with TEx primary phenotype and colored based on the size of each TCR clonotype. Thresholds: average score values across all CD8+ TILs. (C) The proportion of T cells with high memory score is quantified for each R (circles) or NR (diamonds), with lines connecting matched pre-post evaluations. Analysis was performed among all TCR clones (top) or expanded (>2 cells) TCR clones (bottom) with TEx primary phenotype (red) or within TNExM clones (blue) as positive control (Data File S9). The lack of relevant differences in Rs and NRs documents that clonal revival is not significant in this clinical setting. (D,E,F) Quantification of ZNF683+ cytotoxic (CTX+) TEx cells among CD8+ TILs in HNSCC. (D): Bidimensional plot quantifying the expression of ZNF683 (x axis) and cytotoxicity genes (summarized in a score, y axis, see Materials and Methods) in CD8+ TILs with TEx-TCR clonotypes, colored according to their expansion. Thresholds represents average values of variables, as measured in the entire dataset of CD8+ TILs. For each R and NR, the frequencies ZNF683+CTX+ TILs are reported in (E), among all (top) or expanded (>2 cells, bottom) CD8+ TCR clonotypes with TEx primary phenotypes (Data File S9) or among TNexM clones as negative control (blue). (F) Quantification of ZNF683+CTX+ single cells across the overall CD8+ TILs. Bars: mean percentages with SEM. ZNF683+CTX+ TILs are shown with full colors amongst cell states classified as exhausted (red) or memory (blue). Grey: unclassified cells. Analysis was repeated for CD8+ TILs with any TCRs (top) or with expanded (>2 cells) specificities (bottom) (Data File S9). P values: significant comparisons calculated with two-tailed unpaired t-test (R vs NR) or with two-tailed paired ratio t-test (pre vs post).