Abstract
Transcriptional factor RFX6 is known to be a causal gene of Mitchell–Riley syndrome (MRS), an autosomal recessive neonatal diabetes associated with pancreatic hypoplasia and intestinal atresia/malformation. The morphological defects are limited to posterior foregut and mid-hindgut endodermal lineages and do not occur in the anterior foregut lineage; the mechanism remains to be fully elucidated. In this study, we generated RFX6+/eGFP heterozygous knockin and RFX6eGFP/eGFP homozygous knockin/knockout human-induced pluripotent stem cell (hiPSC) lines and performed in vitro endoderm differentiation to clarify the role of RFX6 in early endoderm development. RFX6 expression was found to surge at the primitive gut tube (PGT) stage in comparison with that in the undifferentiated or definitive endoderm stage. At the PGT stage, the expression of PDX1 and CDX2, posterior foregut and mid-hindgut master regulators, respectively, was decreased by the RFX6 deficit. PDX1+ and CDX2+ cells were mostly green fluorescent protein (GFP)+ in RFX6+/eGFP hiPSCs, but their cell number was markedly decreased in RFX6eGFP/eGFP hiPSCs. The expression of SOX2, an anterior foregut marker, was not affected by the RFX6 deficit. In addition, we found a putative RFX6-binding X-box motif using cap analysis of gene expression-seq and the motif-containing sequences in the enhancer regions of PDX1 and CDX2 bound to RFX6 in vitro. Thus, RFX6 regulates the ParaHox genes PDX1 and CDX2 but does not affect SOX2 in early endodermal differentiation, suggesting that defects in early stage endoderm patterning account for the morphological pathology of MRS.
Significance Statement.
This study describes the role of human RFX6 in the early stage endoderm patterning that underlies the organogenetic disorders seen in Mitchell–Riley syndrome, a human RFX6-deficit disease with pancreatic hypoplasia and intestinal atresia/malformation presenting with neonatal diabetes and severe malnutrition. We demonstrate that RFX6 regulates PDX1 and CDX2, posterior foregut and mid-hindgut master transcriptional factors, respectively, but does not affect SOX2, an anterior foregut marker, at the primitive gut tube stage, by using in vitro endoderm differentiation of RFX6+/eGFP heterozygous knockin and RFX6eGFP/eGFP homozygous knockin/knockout human-induced pluripotent stem cell (hiPSC) reporter lines and nongene-modified hiPSCs. In addition, the RFX6-eGFP knockin hiPSC lines generated in this study may contribute to future pancreatic and intestinal endocrine research.
Introduction
The gene of human regulatory factor 6 (RFX6), a transcriptional factor composed of 928 amino acids, encoded by 19 exons on chromosome 6q22, was shown to be a regulatory factor X (RFX) family member (1, 2). RFX6 was then identified as the causative gene of Mitchell–Riley syndrome (MRS; OMIM #615710) (3), which had been clinically recognized as an autosomal recessive disease characterized by endodermal organ dysgenesis including hypoplastic or annular pancreas with neonatal diabetes, intestinal atresia, and gallbladder hypoplasia or aplasia (4–6). Similar human cases with various mutations of RFX6 were reported later (7–12); ectopic gastric tissue in the small intestine was observed in some cases (11, 12). The severity of the symptoms of MRS varies among patients but is often debilitating or fatal and includes severe malnutrition with diarrhea and high mortality in infancy. The pathological and clinical features of the disease suggest defects in the mechanism by which RFX6 regulates tissue patterning and organogenesis of the endoderm.
In regard to the role of Rfx6 in the early developmental stage, a lineage-tracing mouse study revealed that the progeny cells of Rfx6-expressing cells were broadly observed in each endoderm lineage at embryonic day 10.5 but not in other lineages such as ectoderm and mesoderm (3). The study suggests that Rfx6 first appears in the earlier endoderm stage and expresses only in that lineage.
Compared with its broad distribution at the early endoderm stage, RFX6 expression is restricted to the pancreas, small intestine, and colon, which are the posterior foregut and mid-hindgut-derived organs in human adults (13). Particularly in the pancreatic islet and intestinal enteroendocrine cell developmental stage, Rfx6 is considered to be downstream of Ngn3 (Neurogenin 3) and acts as the chief regulator of pancreatic and intestinal endocrine progenitor cell differentiation. Ngn3 knockout mice show no Rfx6 expression in the pancreatic islets (14). NGN3−/− and RFX6−/− human embryonic stem cells (hESCs) also exhibit very few C-peptide-positive cells in artificially differentiated islet-like cells, with almost no expression of INS, GCG, SST, or GHRL (15). Tamoxifen-induced Villin-Cre Rfx6fl/fl, intestine-specific Rfx6 knockout mice, almost completely lack the expression of Gcg, Ghrl, Gip, Pyy, and Cck and partially lack Nts, Sst, and Sct in the intestine (16), while Villin-Cre Ngn3fl/fl mice lack all types of enteroendocrine cells (17). CRISPR-Cas9 mediated ex vivo Rfx6-knockout intestinal organoids of Rosa26-Cas9 mice show the reduction of Gcg, Ghrl, Gip, Cck, and Tph1 expression, while Ngn3 expression is not decreased (18).
However, congenital systemic RFX6 deficit results in more severe morphological disorders than NGN3 deficit. Unlike the severe disorganization of the pancreas and intestine found in MRS, loss-of-function mutation of human NGN3 did not affect the upper gastrointestinal or small intestine follow-through and organ formation (19). Rfx6−/− mice exhibit hypogenesis of the pancreas and digestive tract disorders and therefore fail to feed normally, dying within 2 days postpartum (3), which is not the case in Ngn3−/− mice (20). The morphological discrepancy between the functional deficit of RFX6 and NGN3 suggests another RFX6-active phase in early endoderm development that controls the patterning of endodermal lineages differently from that of the NGN3-RFX6 upstream–downstream relationship operant in the later pancreas and intestinal endocrine cell developmental stage.
This study focuses on the functional role of RFX6 in the early stage of human endoderm patterning and development, which links the genetic features and the morphological disorders of MRS. We used human-induced pluripotent stem cells (hiPSCs), which enables us to recapitulate the early phase of human ontogeny with the least ethical limitations (21). In this study, we approach this unknown mechanism using in vitro endoderm differentiation of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP RFX6-eGFP knockin hiPSC reporter lines that we generated.
Results
Generation of RFX6+/eGFP and RFX6eGFP/eGFP hiPSC lines
We first generated the RFX6+/eGFP heterozygous knockin and RFX6eGFP/eGFP homozygous knockout and knockin cell lines (Fig. 1a). In developing these cell lines, we performed homologous recombination using the bacterial artificial chromosome (BAC) with gene-modified human RFX6, in which an eGFP cassette is inserted just after the 5′ untranslated region (Fig. 1b). The specificity of the knockin locus was confirmed by fluorescent in situ hybridization (FISH) on the knockin reporter gene cassette. Signals of the inserted sequence were precisely identified in the decent target locus, chromosome 6q22, in one allele in RFX6+/eGFP and in both alleles in RFX6eGFP/eGFP. Both cell lines maintain a normal karyotype identical to that in the parental line, 46 XX (Fig. 1c). The undifferentiated status of all cell lines was validated by alkaline phosphatase (ALP) activity and the undifferentiation markers, SSEA4, TRA-1-60, and NANOG (Fig. 1d). Pluripotency was confirmed by all three germ-layer markers, the ectoderm marker β3-Tubulin, the mesoderm marker α-smooth muscle actin (SMA), and the endoderm marker FOXA2, through in vitro differentiation after embryoid body (EB) formation (Fig. 1e and f). Each cell line survived well over 50 passages.
Fig. 1.
The generation of RFX6-eGFP knockin cell lines. a) A schema of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs. b) Schematic procedures to generate RFX6+/eGFP and RFX6eGFP/eGFP hiPSCs. c) A locus validation of RFX6+/eGFP and RFX6eGFP/eGFP hiPSCs by FISH. (Left) Ideogram and Q-banding images of chromosome 6. (Middle) FISH images of chromosome 6. The white triangles mark the signals of the inserted gene. (Right) Karyotype images. d) ALP activity and immunofluorescent images of SSEA4, TRA-1-60, and NANOG of the undifferentiated colony of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs. Bar: 100 µm. e) Bright-field images of in vitro differentiation of EBs. Bar: 1 cm. f) Immunofluorescent images of the ectoderm marker β3-Tubulin, the mesoderm marker α-SMA, and the endoderm marker FOXA2 after in vitro EB differentiation of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs. Bar: 100 µm.
RFX6 is prominently expressed in the primitive gut tube stage
To determine the stage in which the RFX6 expression surges, we differentiated the RFX6+/+ hiPSC parental line to primitive gut tube (PGT) cells in vitro with a slight modification of the previous protocol (22), by which the cells were differentiated into the PGT stage through the definitive endoderm (DE) stage, expressing decent marker genes at each stage (Fig. 2a–d). RFX6 expression was clearly increased at the PGT stage compared with that at the undifferentiated and DE stage (Fig. 2e). We then differentiated RFX6+/eGFP and RFX6eGFP/eGFP hiPSCs into the PGT stage by the same differentiation method to verify reporter gene expression. The general endoderm marker FOXA2 expression was not altered in the three cell lines (Fig. 2f). RFX6 expression decreased in an allele-dependent manner (Fig. 2g) and that of eGFP was detected only in knockin cell lines (Fig. 2h). We obtained similar results by western blotting (Fig. 2i). Immunocytochemistry images show that green fluorescent protein (GFP) was positive only for knockin cell lines and was negative for the parental cell line (Fig. 2j). Coimmunostaining of GFP and RFX6 of the RFX6+/eGFP hiPS line at the PGT stage (day 10) revealed similar signals of GFP and RFX6 but a relatively high nonspecific signal of anti-RFX6 antibody compared with that of anti-GFP antibody. Western blotting data support the immunocytochemistry data (Fig. S1).
Fig. 2.
RFX6 expression during in vitro differentiation to the PGT stage, reporter gene validation, and RNA-seq to search candidate downstream genes of RFX6. a) In vitro differentiation method into the PGT stage (day 10) through the DE stage (day 4). b–e) Marker gene expression and RFX6 expression of RFX6+/+ with the qPCR at each in vitro differentiation stage. (Endogenous control is RPL27. n = 3. Bars are mean ± SD. ns, not significant. *<0.05, **<0.01, ***<0.001 for P-value.) f–h) FOXA2, RFX6, and reporter gene EGFP expression with the qPCR of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage. (Endogenous control is RPL27. n = 3. Bars are mean ± SD. n.d., not detected. ns, not significant. *<0.05, **<0.01, ***<0.001 for P-value.) i) Western blotting images of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage using ACTB as an endogenous control. j) GFP immunofluorescent images of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage. Bar: 100 µm. k) CAGE-seq gene plots comparing RFX6+/+ hiPSCs with RFX6eGFP/eGFP hiPSCs at the PGT stage. The vertical line corresponds to fold change of the adjusted read number; the horizontal line correlates with the adjusted read number itself. The dots of down-regulated and up-regulated genes in RFX6eGFP/eGFP hiPSCs were highlighted respectively. l) A down-regulated gene list of RFX6eGFP/eGFP hiPSCs compared with RFX6+/+ hiPSCs at the PGT stage.
Selection of candidate downstream genes of RFX6
To select candidate genes regulated by RFX6 during the DE stage to the PGT stage, we compared bulk gene expression between RFX6+/+ and RFX6eGFP/eGFP hiPSCs at the PGT stage using the cap analysis of gene expression (CAGE)-seq. Among down-regulated genes in RFX6eGFP/eGFP hiPSCs, expression of the posterior foregut marker PDX1 and the mid-hindgut marker CDX2, both of which are master transcriptional factors in the endoderm development, was detected (23). NGN3, which is thought to be upstream of RFX6 in pancreatic endocrine cells, was also decreased at this stage (Fig. 2k and l).
PDX1 and CDX2 expressing cells are decreased by the RFX6 deficit at the PGT stage
CDX2 expression was significantly up-regulated at the PGT stage compared with that of the predifferentiation and DE stage, similarly to PDX1 (Figs. 2d and 3a). RFX6 expression precedes PDX1 and CDX2 expression from the DE stage to the PGT stage (Fig. S2). Bulk expression of PDX1 and CDX2 at the PGT stage of RFX6eGFP/eGFP hiPSCs was decreased compared with that of RFX6+/+ and RFX6+/eGFP hiPSCs (Fig. 3b and c). A flow cytometry analysis at this stage revealed that 97.1% of PDX1+ cells and 94.1% of CDX2+ cells were GFP+ in RFX6+/eGFP hiPSCs. The proportion of GFP+PDX1+ cells in the GFP+ cells was 60.7% in RFX6+/eGFP hiPSCs, which decreased to 13.8% in RFX6eGFP/eGFP; that of GFP+CDX2+ cells in GFP+ cells was 76.0% in RFX6+/eGFP and 33.7% in RFX6eGFP/eGFP hiPSCs (Fig. 3d and e). Immunocytochemistry images indicated that GFP+PDX1+ cells were more predominantly observed than GFP−PDX1+ cells in RFX6+/eGFP hiPSCs, and the number of GFP+PDX1+ cells was decreased in RFX6 eGFP/eGFP hiPSCs compared with that in RFX6 +/eGFP hiPSCs, bolstering the results of the flow cytometry analysis (Fig. 3f). In regard to the PDX1 and CDX2 relationship, PDX1/CDX2 costaining shows that PDX1 and CDX2 are expressed simultaneously in some PGT cells in addition to PDX1+CDX2− and PDX1−CDX2+ cells in RFX6+/eGFP hiPSCs, although the number of PDX1+CDX2+ cells was not as large as that of PDX1+CDX2− and PDX1−CDX2+ cells (Fig. 3g). The number of CDX2+ cells was decreased in RFX6eGFP/eGFP hiPSCs compared with that in RFX6+/+ and RFX6+/eGFP hiPSCs (Fig. 3g). Multiple clones of RFX6+/eGFP and RFX6eGFP/eGFP hiPSC lines were assessed for PDX1 and CDX2 expression (Fig. S3). We compared another PDX1-prone differentiation protocol with the same differentiation duration (24). While CDX2 expression remained relatively lower than the protocol used in this study, a similar decrease in PDX1 and CDX2 expressions due to the RFX6 deficit was nevertheless observed (Fig. S4).
Fig. 3.
RFX6 regulates the posterior foregut marker PDX1 and mid-hindgut marker CDX2 at the PGT stage. a) CDX2 expression of RFX6+/+ hiPSCs at each in vitro differentiation stage (n = 3). b and c) PDX1 and CDX2 expression of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage. (n = 6. Endogenous control is RPL27. Bars are mean ± SD. ns, not significant. ***<0.001, ****<0.0001 for P-value.) d and e) A flow cytometry analysis of PDX1/GFP and CDX2/GFP at the PGT stage. GFP was unstained only with CDX2 due to antibody cross-activity. Most PDX1+ and CDX2+ cells are GFP+, but PDX1+GFP+ and CDX2+GFP+ cell populations are reduced in RFX6eGFP/eGFP hiPSCs compared with those in RFX6+/eGFP hiPSCs. f and g) Immunofluorescent images of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage. Bar: 100 µm. f) PDX1 and GFP costaining images. In RFX6+/eGFP hiPSCs, PDX1+GFP+ cells are more dominant than PDX1+GFP− cells. Such PDX1+GFP+ cells are sparse in RFX6eGFP/eGFP hiPSCs compared with the number in RFX6+/eGFP hiPSCs. g) PDX1 and CDX2 costaining images. Some PDX1+CDX2+ cells in addition to PDX1+CDX2− and PDX1−CDX2+ cells are seen in RFX6+/+ and RFX6+/eGFP hiPSCs. PDX1+ and CDX2+ cells are observed sporadically in RFX6eGFP/eGFP hiPSCs.
The expression of SOX2, an anterior foregut marker, was not affected by the RFX6 deficit
The expression of SOX2, which is known as an anterior foregut marker and an undifferentiation marker, was significantly decreased from the undifferentiation stage to the DE stage and tended to increase from DE to the PGT stage (Fig. 4a). At the PGT stage, SOX2 expression did not differ among RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs (Fig. 4b). Flowcytometry analysis of SOX2 and GFP at the PGT stage indicated that the majority of SOX2+ cells were GFP− in both RFX6+/eGFP and RFX6eGFP/eGFP hiPSCs (Fig. 4c). Immunocytochemistry images confirmed this finding (Fig. 4d).
Fig. 4.
RFX6 does not down-regulate the anterior foregut marker SOX2. a) SOX2 expression of RFX6+/+ hiPSCs at each in vitro differentiation stage. b) SOX2 expression of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage. (Endogenous control is RPL27. n = 3. Bars are mean ± SD. ns, not significant. **<0.01, ***<0.001 for P-value.) c) A flow cytometry analysis of SOX2/GFP at the PGT stage. Most SOX2+ cells are GFP− and vice versa, in both RFX6+/eGFP and RFX6eGFP/eGFP hiPSCs. d) SOX2/GFP immunofluorescent images of RFX6+/+, RFX6+/eGFP, and RFX6eGFP/eGFP hiPSCs at the PGT stage.
A CATGGCAA X-box motif was detected using CAGE-seq as a target of RFX6
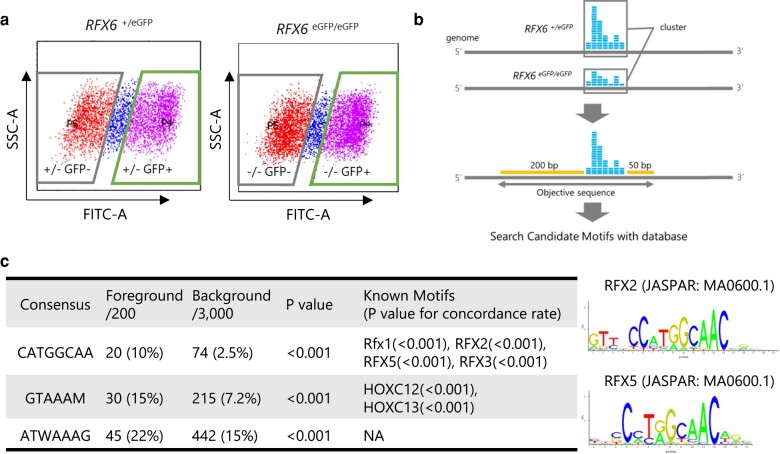
We also searched for a candidate RFX6-binding motif sequence by comparing GFP-positive cells of RFX6+/eGFP hiPSCs with those of RFX6eGFP/eGFP hiPSCs at the PGT stage by using CAGE-seq (Figs. 5a, b, and S5). By using motif databases, putative RFX6-binding motifs were surveyed in the sequencing read cluster in the range from 200 bp upstream to 50 bp downstream of the transcriptional starting site (TSS). The top 200 clusters down-regulated in RFX6eGFP/eGFP hiPSCs in comparison with RFX6+/eGFP hiPSCs were selected; the sequences of these clusters were compared with 3,000 random clusters that have no difference in both groups (Fig. 5b). Among the candidate motifs, we found a CATGGCAA motif that has not been reported previously as an RFX6-binding motif but is known to be an X-box motif of other RFX family members such as RFX2, RFX3, and RFX5 (Fig. 5c).
Fig. 5.
A RFX6-binding motif search. a) A living cell flow cytometry of RFX6+/eGFP and RFX6eGFP/eGFP hiPSCs at the PGT stage. GFP-positive, GFP-negative, and intermediate cells are highlighted separately. SSC-A, side scatter area; FITC-A, fluorescein isothiocyanate area. b) A schema of the motif search using CAGE-seq and motif databases. c) (left) Putative consensus motifs found by CAGE-seq and motif databases. Foreground objective sequences represent read clusters less frequently observed in RFX6eGFP/eGFP hiPSCs than in RFX6+/eGFP hiPSCs. Background sequences are those that did not show differences. Motif frequency in both foreground and background is also described as percentage. The P-value in the independent column denotes the difference in the frequency between foreground and background. The P-value in the parenthesis is calculated by using the concordance rate to the known motifs. (right) RFX2 and RFX5 ChIP-seq motif logos from the JASPAR database.
RFX6 binds to the CATGGCAA X-box site upstream of the PDX1 and CDX2 genes on the electrophoresis mobility shift assay
In the sequence of PDX1 and CDX2 genes, which are located closely at 13q12.2, the CATGGCAA motif was detected 10 kb upstream and 26 kb upstream of the TSS, respectively. The binding capacity of RFX6 to the motif was confirmed by electrophoresis mobility shift assay (EMSA) using a nonradioisotope method with biotin-end-labeled oligos. Purified RFX6 protein from HEK293-EF1α-RFX6-C-Myc-IRES-Puro transgenic cells was applied to EMSA. RFX6 protein bound to the labeled CATGGCAA-containing sequences found upstream of PDX1 and CDX2; the binding was cancelled out by the excess nonbiotin-labeled probes (Fig. 6a). Signals of these bindings to each target sequence were not detected when the probe sequences lacked the CATGGCAA X-box (Fig. 6b).
Fig. 6.
RFX6 and putative X-box motif binding in a non-RI EMSA. a) EMSA images of the RFX6 protein and the PDX1-upstream or CDX2-upstream CATGGCAA motif sequence. The shift band diminishes by the excess amount of the nonbiotinylated probes. b) EMSA images comparing the binding capacity of the X-box-containing probe with the X-box-deleted probe to the RFX6 protein. The shift band is not detected with the X-box-deleted probe.
Discussion
This study highlights the function of human RFX6 in the early endoderm developmental stage using eGFP reporter iPSC lines and provides clues to the relationship between the genetic and pathological features of MRS. The RFX6 expression level is lower in the early developmental stages, making it more difficult to quantify its expression level with its antibody than it is in the adult/mature stage. Even so, eGFP, a highly sensitive reporter gene, permitted fine monitoring of the RFX6 expression pattern, by which we found the clue to the pathophysiology of MRS: RFX6 positively affects PDX1 and CDX2 but not SOX2 at the PGT stage (Fig. 7).
Fig. 7.
The predicted pathophysiology of MRS. A schematic figure of the predicted pathophysiology of MRS. RFX6 deficit causes PDX1 and CDX2 down-regulation, while not affecting SOX2. Patients with MRS, therefore, present with only the malformation of the posterior foregut–derived organs and mid-hind gut-derived organs but not the anterior foregut-derived organs.
Thus, the malformation of the pancreas and intestine, and severe diarrhea, the most frequent symptoms of MRS, can be ascribed to down-regulated PDX1 and CDX2 expression at the PGT stage, in which almost all PDX1+ and CDX2+ cells are RFX6+ and the number of PDX1+ cells and CDX2+ cells is decreased by the RFX6 deficit. Pdx1 is the master regulator in pancreas organogenesis (25, 26) and also maintains gastroduodenal order in adults (27). Embryonic Cdx2 expression begins at E8.5 in the posterior gut (28); systemic Cdx2-null mice die around E3.5−5.5 (29) due to Cdx2 expression in the trophectoderm at E3.5 (28). Villin-Cre/Cdx2fl/fl intestinal Cdx2 knockout mice exhibit hypogenesis and loss of polarity of the small intestine at E18.5 (30). PDX1 and CDX2 were found to be partly coexpressed at the PGT stage in our study, as in a previous report alluding to a Pdx1+/Cdx2+ mixed region (27).
Contrary to the effect of the RFX6 deficit on PDX1 and CDX2, the deficit did not significantly affect the anterior foregut marker SOX2. In regard to expression patterning at the PGT stage, interestingly, almost all of the differentiated cells at that stage were SOX2+ or RFX6+ and were distinctly segregated and complementary from each other, nearly all PDX1+ cells and CDX2+ cells being RFX6+. Interestingly, ectopic gastric tissue in the small intestine, which is reported in some patients with MRS (11, 12), may be regarded as another consequence of the patterning disruption. It is known that the Pdx1 deficit results in anteriorization of the Pdx1-positive intestinal domain (27). Thus, the aberrant gastric tissue found in the small intestine of patients with MRS may well result from disruption by the RFX6 deficit, in which SOX2-dominant anterior foregut tube-prone tissue partially supplants the region destined by RFX6 to differentiate into posterior foregut and mid-hindgut-derived tissue. Meanwhile, possible change in the direction of differentiation toward gastric specification by the RFX6 deficit was not detected at the PGT stage in the comparison between RFX6eGFP/eGFP and RFX6+/+ hiPSCs, nor was hepatic specification (Fig. S6). Gene expression alterations of histone acetyltransferase EP300, which has a promotive effect on liver development, and histone methyltransferase EZH2, which has a suppressive effect on pancreas development, were also not observed (31) (Fig. S6). Precise characterization will be a future consideration.
From a clinical viewpoint, ectopic gastric mucosa providing gastric acid secretion leads to pH changes in the small intestine and can cause diarrhea and intestinal ulcer. While only a few reports have surveyed the ectopic gastric mucosa, some of them have reported severe diarrhea and intestinal ulcer (11, 12) and, surprisingly, the resection of the ectopic gastric mucosa dramatically alleviates these symptoms in some cases (11). Thus, an intensive survey and a similar approach might be profitably applied in other MRS cases.
In regard to the direct binding of RFX6 to DNA, the CATGGCAA motif found in our study has been validated in several other members of the human RFX family, including RFX2, RFX3, and RFX5, although they do not have an entirely identical sequence of a DNA-binding domain (1). CCA(G/T)GT(C/T)(C/A)T, identified as the putative motif by the mouse ChIP assay using anti-mouse Rfx6 antibody (32), did not appear in our study sample. ChIP-validated RFX2 (MA0600.1) and RFX5 (MA0510.1) motifs have a robust frequency from −500 to +500 bp of the TSS within the range of −5,000 to +2,000 bp of the TSS (33), but it remains to be elucidated where these motifs are functional outside that range, including in the putative enhancer regions. The CATGGCAA motif, although not found in the PDX1 and CDX2 promoter regions, was found 10 kb upstream of the PDX1 TSS and 26 kb upstream of the CDX2 TSS and bound to RFX6. Interestingly, the PDX1 and CDX2 genes are located face-to-face on human chromosome 13. As similar functional gene positions are close to each other (34) and some adjacent genes share regulation mechanisms (35), the direct binding of RFX6 and the upstream X-box region of PDX1/CDX2 genes suggests a possible function as an enhancer region.
The finding that the expression of NGN3, which is known as an upstream of RFX6 in pancreatic endocrine cells, was decreased at the PGT stage further suggests the multiphasic role of RFX6 in addition to the roles described in later developmental stages, including those as a developmental regulator in pancreatic islets and intestinal endocrine cells and a secretory regulator of mature pancreatic beta-cells (36). As the concept of multiphasic expression of transcriptional factors was reported previously (37), it is helpful to understand why the NGN3-RFX6 upstream–downstream relationship in the later pancreatic and intestinal endocrine cell development and maturation stages cannot satisfactorily explain the morphological discrepancy between the NGN3-null and RFX6-null phenotypes.
Our RFX6 iPSC reporter lines will contribute to future studies regarding the secretory machinery of pancreatic beta cells and enteroendocrine cells. Pancreatic-endocrine-specific Rfx6-deficiency mice show that Rfx6 regulates genes that are involved in the insulin secretion (36); RFX6-knockdown of the human beta-cell line EndoC-βH2 (38) demonstrates the down-regulation of potassium channel genes (39). In addition, Rfx6 up-regulates gastric inhibitory peptide (GIP) expression in enteroendocrine K cells in mice (40). It has also been found that some patients with maturity-onset diabetes of the young have an RFX6 heterozygous-deficient genotype and present a decrease in both fasting and stimulated GIP secretion in the oral glucose tolerance test (41).
Additionally, among the up-regulated genes in the RFX6 KO line (Fig. S6, related to Fig. 2k), CPB1 encodes carboxypeptidase B1 and works specifically in pancreatic exocrine cells. CPB1 up-regulation may reflect the inhibition of endocrine cell differentiation by RFX6 deficit. Although we mainly focused on the master regulator genes of the pancreas and intestine, influenced organs by the RFX6 deficit, such as PDX1 and CDX2 in this study, these functionally essential genes will be a promising target in future studies.
The findings of this study must be interpreted in consideration of some limitations. The disruption of endoderm regulation and patterning by the RFX6 deficit has not been validated in in vivo human RFX6-null embryo or fetus. Future studies are required to determine whether and how the boundary between SOX2+ cells and RFX6+ cells observed in the present study forms and what factors determine the boundary. In regard to RFX6 regulation of PDX1 and CDX2, although the binding of RFX6 and the CATGGCAA X-box was confirmed, it remains to be seen whether RFX6 regulates them directly or indirectly.
Our findings indicate that human RFX6 is essential in endoderm patterning at the PGT stage, regulating the posterior foregut marker PDX1 and mid-hindgut marker CDX2 but not the anterior foregut marker SOX2.
Methods
hiPSC parental line
hiPSC 409B2, a transgene- and virus-free iPSC generated from a Caucasian female, was used as a parental line (42).
Feeder-free hiPSC culture
About 60–70% confluent cells were detached with 0.5 mM ethylenediaminetetraacetic acid (EDTA)/phosphate-buffered saline (PBS) for 10 min at 37 °C. After adding AK02N (Ajinomoto) with 10 μM Y-27632 and centrifuging at 300×g for 5 min, the cells were seeded on iMatrix-511 silk (Nippi)-coated wells with AK02N with 10 μM Y-27632 with a split ratio of 1:200–1:500. Precoating and medium changes were performed according to the manufacturer's protocol.
Human RFX6-BAC recombineering
We used the homologous recombination method (43). First, a human RFX6 BAC clone RP11-451L17 was transformed into Escherichia coli DH10B. Next, 50 bp 5′- and 3′-homology primer arms, identical sequences just before and after the first ATG of the human RFX6 gene, were attached to the eGFP-loxP-PGK-NeoR-loxP cassette by PCR (98 °C 30 s, 30 cycles of 98 °C 10 s, 55 °C 30 s, and 72 °C 90 s, and 72 °C 10 min). Red/ET plasmid pSC101-BAD-gbaA was introduced to hRFX6 BAC-containing E. coli by electroporation, and the knockin cassette was introduced by electroporation (2,500 V, 25 µF, and 200 Ω in 2 mm cuvette). After the G418 antibiotic selection and BAC purification, the recombination was confirmed by Sanger sequencing.
Generation of RFX6-GFP knockin hiPSC lines
A 30-µg knockin cassette-containing hRFX6 BAC was linearized with PI-SceI and introduced to ∼1.5 × 106 parental hiPSCs by electroporation. Cells were cultured in AK02N with 10 μM Y-27632 on day 0, AK02N without Y-27632 on day 1, and AK02N with 100 ng/µL G418 for about 2 weeks afterwards. We selected 15 candidate clones from 90 resistant clones by qPCR of genomic DNA.
Next, we removed the floxed NeoR lesion by transient Cre expression by using pHAGE2-EF1α-Cre-IRES-Puro (Addgene) (44). Approximately, 1 × 106 undifferentiated RFX6+/eGFP-loxP-PGK-NeoR-loxP cells were incubated with the mixture containing 90 μL Opti-MEM (Thermofisher) with 8 μL FuGENE HD (Promega) and 2 μg pHAGE2-EF1α-Cre-IRES-PuroR and then added to 6 mL AK02N with 10 μM Y-27632 and seeded on three wells of an iMatrix 511-silk precoated six-well plate. Cells were cultured in AK02N with 1 μg/mL Puromycin and without Y-27632 on the day after lipofection (day 1), and the medium changes were done with AK02N without Puromycin and Y-27632 every other day from day 3. We picked up candidate colonies after 7–10 days. We selected the colony in which neither NeoR nor Cre was detected in genomic DNA as RFX6+/eGFP-(NeoR deleted) (RFX6+/eGFP) hiPSCs. Finally, we repeated the same procedure of electroporation of the linearized BAC with the RFX6+/eGFP-(NeoR deleted) clone, G418 selection, and PCR confirmation and generated RFX6eGFP-(NeoR deleted)/eGFP-loxP-PGK-NeoR-loxP (RFX6eGFP/eGFP) hiPSCs.
Q-banding karyotype and FISH
Q-banding karyotype analysis and FISH were performed at Chromosome Science Lab Inc. (Hokkaido, Japan). Q-banding analysis was performed on preparations processed with a Hoechst 33258 and quinacrine mustard double staining technique (45). For FISH, probe DNA plasmid, pBS II-EcoRV-eGFP-pA-PGK-NeoR-pA, generated from pBluescript II KS(-) (Agilent) was labeled with Cy3 by nick translation. After hybridization, stringency wash, and counterstaining with DAPI, the probe signal was detected on a Leica CW-4000 cytogenetic workstation.
Immunofluorescent staining
Cells were fixed with 4% paraformaldehyde. Except for SSEA4, cells were permeabilized with 100% ethanol at −20 °C for 10 min. After washing with PBS three times and blocking with Protein Block Serum-Free (PBSF, Dako/Agilent) at room temperature for 1 h, primary antibodies with PBSF were applied at 4 °C overnight. After washing with PBS three times, secondary antibodies with PBSF were applied at room temperature for 1 h. After washing with PBS three times, the cells were mounted with 5× PBS-diluted DAPI-Fluoromount G (SouthernBiotech). Fluorescence microscope images were captured with BZ-X710 (KEYENCE) or Dragonfly (Andor/Oxford Instruments). Antibodies are listed in Table S2.
ALP activity staining
BCIP/NBT Substrate System (Dako) was used according to the manufacturer's instruction.
EB formation and in vitro trilineage differentiation
Undifferentiated hiPSC colonies were detached and applied to the floating culture with AK02N with 10 µM Y-276342 on day 0 and with only AK02N afterwards. After sphere-shaped EB formation and expansion for 5–7 days, EBs were put on the gelatin-coated dish for the following week and harvested with DMEM with 10% FBS to form trilineage cells spontaneously. Trilineage differentiation was confirmed by immunocytochemistry of the ectoderm marker β3-Tubulin, the mesoderm marker α-SMA, and the endoderm marker FOXA2.
Differentiation from hiPSC to PGT cells
Subconfluent hiPS colonies were detached, and 200,000 iPS cells/well were seeded on the iMatrix-precoated well of a 24-well plate with 1 mL AK02N with 10 μM Y-276342 on day 0. Cells were cultured with 1 mL RPMI1640 with 2% vol/vol B27 supplement, 100 ng/mL activin A (DE basal medium), and 3 μM CHIR99021 on day 1, DE basal medium with 1 μM CHIR99021 on day 2, and DE basal medium only on day 3 followed by 1 mL improved MEM Zinc Option medium with 1% B27, 1 μM dorsomorphin, 2 μM retinoic acid, and 10 μM SB431542 on days 4, 6, and 8. Medium changes were made every 24 ± 2 h until day 4 and every 48 ± 2 h afterwards. We also used another PDX1-prone differentiation protocol to validate the consistency of the major results (Fig. S4) (24).
RNA extraction and qPCR
We used RNA-easy Kit, QIAshredder, and RNase-Free DNase Set (Qiagen, USA) for RNA extraction, ReverTra Ace qPCR RT Kit (Toyobo, Japan) for reverse transcription, and Thunderbird SYBR qPCR Mix for qPCR (Toyobo), according to the manufacturer's protocols. The amount of cDNA used for qPCR was equivalent to 40 ng total RNA. qPCR was performed at 95 °C for 10 min and 40 cycles at 95 °C for 15 s and at 60 °C for 60 s on StepOne Plus (Applied Biosystems, USA). 2ΔCt value was used in the data analysis. Primers are listed in Table S1.
Flow cytometry
For immunofluorescent staining, detached cells were pipetted with fixation/permeabilization solution (BD) in the ratio of 1 μL/1 × 104 cells and incubated at room temperature for 30 min. After centrifuging at 300×g for 5 min, pellet cells were mixed with 2% donkey serum (DS) in 1× perm/wash (PW) buffer (BD) in the ratio of 1 mL/2 × 106 cells and stored at 4 °C. Fifty microliters of the mixture were applied onto the 96-well plate. After centrifuging at 400×g for 3 min (*), pellet cells and the primary antibody diluted with 100 μL 2% DS-PW were mixed at 4 °C overnight. After repeating (*), pellet cells were washed with PW buffer twice. Pellet cells and secondary antibodies diluted with 100 μL 2% DS-PW were mixed at room temperature for 1 h. After washing with PW buffer twice, the cells were sorted by FACS Aria 2 (BD). In flow cytometry of CDX2 and GFP, CDX2 was stained and GFP was unstained. Antibodies are listed in Table S2.
Cap analysis of gene expression-seq
CAGE library preparation, sequencing, mapping, gene expression, and motif discovery analysis were performed by DNAFORM (Kanagawa, Japan). cDNA was synthesized from total RNA using random primers. Ribose diols in the 5′-cap structures of RNA were oxidized and biotinylated. The biotinylated RNA/cDNAs were selected by streptavidin beads after RNase digestion. After an adaptor ligation to both cDNA ends, double-stranded cDNA libraries were sequenced using single-end 75-nucleotide reads on a NextSeq 500 instrument (Illumina). CAGE-tag reads were mapped to the RefSeq GRCh38 using BWA (ver. 0.5.9). Unmapped reads were mapped by HISAT 2 (ver. 2.0.5). For the bulk CAGE experiment, CAGE-tag clustering and analysis were performed by CAGEr (46). Genes with Log2-fold change below −2.0 or above +2.0 and base means above 50 are listed, and WebGestalt2019 was used for overrepresentation analysis for these gene lists (47). For motif analysis experiment, pipeline RECLU (48) was used. In this procedure, genomic DNA sequences from 200 bp upstream to 50 bp downstream of differentially expressed CAGE peaks were subjected to de novo motif discovery tools.
Western blotting
Fifty milligrams of whole lysate protein were extracted with radio-immunoprecipitation assay (RIPA) Buffer with 1% Protease Inhibitor (Nacalai). The concentration was calculated by Protein Assay BCA kit (Nacalai). Samples were treated with 6× Sample Buffer Solution with 2-ME at 95 °C for 5 min. Electrophoresis was performed using Criterion Cell, Criterion Blotter, Criterion XT Precast Gel (Bio-Rad), and Immobilon-P PVDF membrane (Merck). Protein separation was carried out at 200 V for 1 h with a 20× DW-diluted XT MOPS Running Buffer (Bio-Rad). Membrane transfer was performed at 120 V for 1 h with the transfer buffer (25 mM Tris-HCl, 192 mM Glycine, and 10% Methanol in DW). We used 4% Block Ace (UKB) diluted with PBS for blocking at room temperature for 1 h. After 2× wash with PBS for 10 min, the primary antibody was applied overnight at 4 °C. Then, after 2× wash with 0.2% Block Ace with PBS (PBS-B) for 10 min, the secondary antibody was applied for 1 h at room temperature. After 4× wash with 0.1% Tween 20 with PBS-B, chemiluminescence reaction was performed using Chemi-Lumi One Super (Nacalai). Blocking, washing, and antibody reactions were done on a plate shaker. Image capture was performed with ImageQuant LAS-4000 mini (GE Healthcare/Cytiva). Antibodies are listed in Table S2.
Electrophoresis mobility shift assay
For protein extraction, HEK293-EF1α-RFX6-Myc-DDK-IRES-Puro overexpression cells were generated by transfection of pLenti-EF1α-hRFX6ORF-Myc-DDK-IRES-Puro (OriGene) to HEK293 cells. Confluent cells of a 10-cm culture plate were detached with 0.25% Trypsin/EDTA and precipitated by centrifuge. After the PBS wash, pellet cells were treated with 500 µL M-PER Mammalian Protein Extraction Reagent (Thermofisher) with 1% Protease Inhibitor. The lysate was purified using c-Myc tagged Protein MILD PURIFICATION KIT ver. 2 (MBL) according to the manufacturer's protocol with slight modification in the rotation time of the beads-lysate mixture from 1 h to overnight. The purified protein concentration was ∼0.5–1 µg/µL. Oligonucleotide probes for EMSA were dissolved with TEN Buffer (10 mM Tris; pH 8.0, 1 mM EDTA, and 0.1 mM NaCl) and annealed in a 1:1 sense–antisense ratio using natural cooling to room temperature after treatment at 94 °C for 10 min.
Protein size separation was performed using Mini-PROTEAN Tetra Cell, 5% Mini-PROTEAN TBE Precast Gel (Bio-Rad), and 1×Tris-borate EDTA (TBE) Buffer. In a 20-µL assay sample, 1.25 µg protein, 40 fmol biotinylated probe, 0–8 pmol nonbiotinylated probe, 2 µL 10× Binding Buffer (100 mM Tris, 500 mM KCl, 10 mM DTT; pH 7.5), 3 µL 50% glycerol, 1 µL 60 mM MgCl2, 1 µL 10 mM EDTA, and DW up to 20 µL were used. After the prerun at 100 V for 45 min and concurrent protein–probe reaction at 25 °C for 20 min, 20 µL of the sample mixture with 2 µL 5 × Nucleic Acid Sample Loading Buffer (Bio-Rad) was electrophoresed at 100 V for 35 min. Membrane transfer was performed using Trans-Blot Turbo, Zeta-Probe Blotting Membranes (Bio-Rad), and 0.5× TBE Buffer. After equilibration with 0.5× TBE for 10 min, transfer was performed at 0.2 A for 1 h. DNA crosslink to the membrane was performed at 120 mJ/cm2 with UV Crosslinker, CL-1000 (UVP). Signal detection of biotin-labeled probes was performed with Chemiluminescent Nucleic Acid Detection Module Kit (Thermofisher). Images were captured with ImageQuant LAS-4000 mini (GE Healthcare/Cytiva).
Data analysis
All comparative analyses for qPCR were performed with one-way ANOVA followed by Tukey's multiple comparison test using GraphPad Prism ver. 9.5.1 (GraphPad Software). P-value <0.05 was regarded as statistically significant.
Supplementary Material
Acknowledgments
The authors thank Sara Yasui, Saki Kanda, Ayaka Shiota, and Yoshie Fukuchi for their laboratory assistance. The authors also thank the Medical Research Support Center, Graduate School of Medicine, Kyoto University, for using their research facility.
Contributor Information
Toshihiro Nakamura, Department of Diabetes, Endocrinology and Nutrition, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan.
Junji Fujikura, Department of Diabetes, Endocrinology and Nutrition, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan.
Ryo Ito, Department of Diabetes, Endocrinology and Nutrition, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan; Center for iPS Cell Research and Application, Kyoto University, Kyoto 606-8507, Japan.
Yamato Keidai, Department of Diabetes, Endocrinology and Nutrition, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan.
Nobuya Inagaki, Department of Diabetes, Endocrinology and Nutrition, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan; Medical Research Institute, Kitano Hospital, PIIF Tazuke-kofukai, Osaka 530-8480, Japan.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
This study was supported by the Japan Society for the Promotion of Science (JSPS KAKENHI grant numbers 21K06970 and 22K08668), the Japan Foundation for Applied Enzymology (Front Runner of Future Diabetes Research), the Foundation of Future Research Support and the Japan Agency for Medical Research and Development (JP23ym0126125). The authors also acknowledge the support of the Japan Association for Diabetes Education and Care through an international travel grant.
Author Contributions
T.N. contributed to the study conception, researched the data, and wrote the manuscript. J.F. contributed to the study conception, researched the data, and revised the manuscript. R.I. researched the data and contributed to the discussion. Y.K. researched the data and contributed to the discussion. N.I. organized the study and contributed to the discussion.
Data Availability
The data that support the findings of this study are available in the Supplementary material.
Ethical Approval
This study was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine and Kyoto University Hospital (R0091).
References
- 1. Aftab S, Semenec L, Chu JS, Chen N. 2008. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 8:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Emery P, Durand B, Mach B, Reith W. 1996. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 24:803–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith SB, et al. 2010. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 463:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mitchell J, et al. 2004. Neonatal diabetes, with hypoplastic pancreas, intestinal atresia and gall bladder hypoplasia: search for the aetiology of a new autosomal recessive syndrome. Diabetologia. 47:2160–2167. [DOI] [PubMed] [Google Scholar]
- 5. Chappell L, et al. 2008. A further example of a distinctive autosomal recessive syndrome comprising neonatal diabetes mellitus, intestinal atresias and gall bladder agenesis. Am J Med Genet Part A. 146:1713–1717. [DOI] [PubMed] [Google Scholar]
- 6. Martinovici D, et al. 2010. Neonatal hemochromatosis and Martinez-Frias syndrome of intestinal atresia and diabetes mellitus in a consanguineous newborn. Eur J Med Genet. 53:25–28. [DOI] [PubMed] [Google Scholar]
- 7. Spiegel R, et al. 2011. Clinical characterization of a newly described neonatal diabetes syndrome caused by RFX6 mutations. Am J Med Genet Part A. 155:2821–2825. [DOI] [PubMed] [Google Scholar]
- 8. Concepcion JP, et al. 2014. Neonatal diabetes, gallbladder agenesis, duodenal atresia, and intestinal malrotation caused by a novel homozygous mutation in RFX6. Pediatr Diabetes. 15:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khan N, Dandan W, Al Hassani N, Hadi S. 2016. A newly-discovered mutation in the RFX6 gene of the rare Mitchell-Riley syndrome. J Clin Res Pediatr Endocrinol. 8:246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zegre Amorim M, et al. 2015. Mitchell-Riley syndrome: a novel mutation in RFX6 gene. Case Rep Genet. 2015:937201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skopkova M, et al. 2016. Two novel RFX6 variants in siblings with Mitchell-Riley syndrome with later diabetes onset and heterotopic gastric mucosa. Eur J Med Genet. 59:429–435. [DOI] [PubMed] [Google Scholar]
- 12. Sansbury FH, et al. 2015. Biallelic RFX6 mutations can cause childhood as well as neonatal onset diabetes mellitus. Eur J Hum Genet. 23:1744–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pearl EJ, Jarikji Z, Horb ME. 2011. Functional analysis of Rfx6 and mutant variants associated with neonatal diabetes. Dev Biol. 351:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soyer J, et al. 2010. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 137:203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu Z, et al. 2016. Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell. 18:755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piccand J, et al. 2019. Rfx6 promotes the differentiation of peptide-secreting enteroendocrine cells while repressing genetic programs controlling serotonin production. Mol Metab. 29:24–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mellitzer G, et al. 2010. Loss of enteroendocrine cells in mice alters lipid absorption and glucose homeostasis and impairs postnatal survival. J Clin Invest. 120:1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gehart H, et al. 2019. Identification of enteroendocrine regulators by real-time single-cell differentiation mapping. Cell. 176:1158–1173.e16. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, et al. 2006. Mutant neurogenin-3 in congenital malabsorptive diarrhea. N Engl J Med. 355:270–280. [DOI] [PubMed] [Google Scholar]
- 20. Gradwohl G, Dierich A, LeMeur M, Guillemot F. 2000. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 97:1607–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takahashi K, et al. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872. [DOI] [PubMed] [Google Scholar]
- 22. Kunisada Y, Tsubooka-Yamazoe N, Shoji M, Hosoya M. 2012. Small molecules induce efficient differentiation into insulin-producing cells from human induced pluripotent stem cells. Stem Cell Res. 8:274–284. [DOI] [PubMed] [Google Scholar]
- 23. Mcgrath PS, Wells JM. 2015. SnapShot: GI tract development. Cell. 161:176.e1. [DOI] [PubMed] [Google Scholar]
- 24. Ito R, et al. 2023. Elucidation of HHEX in pancreatic endoderm differentiation using a human iPSC differentiation model. Sci Rep. 13:8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jonsson J, Carlsson L, Edlund T, Edlund H. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 371:606–609. [DOI] [PubMed] [Google Scholar]
- 26. Offield MF, et al. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 122:983–995. [DOI] [PubMed] [Google Scholar]
- 27. Holland AM, Garcia S, Naselli G, Macdonald RJ, Harrison LC. 2013. The parahox gene Pdx1 is required to maintain positional identity in the adult foregut. Int J Dev Biol. 57:391–398. [DOI] [PubMed] [Google Scholar]
- 28. Beck F, Erler T, Russell A, James R. 1995. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 204:219–227. [DOI] [PubMed] [Google Scholar]
- 29. Chawengsaksophak K, James R, Hammond VE, Köntgen F, Beck F. 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 386:84–87. [DOI] [PubMed] [Google Scholar]
- 30. Grainger S, Savory JGA, Lohnes D. 2010. Cdx2 regulates patterning of the intestinal epithelium. Dev Biol. 339:155–165. [DOI] [PubMed] [Google Scholar]
- 31. Xu CR, et al. 2011. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 332:963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng C, et al. 2019. Identification of Rfx6 target genes involved in pancreas development and insulin translation by ChIP-seq. Biochem Biophys Res Commun. 508:556–562. [DOI] [PubMed] [Google Scholar]
- 33. Sugiaman-Trapman D, et al. 2018. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genomics. 19:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thévenin A, Ein-Dor L, Ozery-Flato M, Shamir R. 2014. Functional gene groups are concentrated within chromosomes, among chromosomes and in the nuclear space of the human genome. Nucleic Acids Res. 42:9854–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imai KS, Daido Y, Kusakabe TG, Satou Y. 2012. Cis-acting transcriptional repression establishes a sharp boundary in chordate embryos. Science. 337:964–967. [DOI] [PubMed] [Google Scholar]
- 36. Piccand J, et al. 2014. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 9:2219–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Villasenor A, Chong DC, Cleaver O. 2008. Biphasic Ngn3 expression in the developing pancreas. Dev Dyn. 237:3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scharfmann R, et al. 2014. Development of a conditionally immortalized human pancreatic β cell line. J Clin Invest. 124:2087–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chandra V, et al. 2014. RFX6 regulates insulin secretion by modulating Ca2+ homeostasis in human β cells. Cell Rep. 9:2206–2218. [DOI] [PubMed] [Google Scholar]
- 40. Suzuki K, et al. 2013. Transcriptional regulatory factor X6 (RFX6) increases gastric inhibitory polypeptide (GIP) expression in enteroendocrine k-cells and is involved in GIP hypersecretion in high fat diet-induced obesity. J Biol Chem. 288:1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel KA, et al. 2017. Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun. 8:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okita K, et al. 2011. A more efficient method to generate integration-free human iPS cells. Nat Methods. 8:409–412. [DOI] [PubMed] [Google Scholar]
- 43. Mae S-I, et al. 2013. Monitoring and robust induction of nephrogenic intermediate mesoderm from human pluripotent stem cells. Nat Commun. 4:1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Somers A, et al. 2010. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 28:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoshida MC, Ikeuchi T, Sasaki M. 1975. Differential staining of parental chromosomes in interspecific cell hybrids with a combined quinacrine and 33258 Hoechst technique. Proc Jpn Acad. 51:184–187. [Google Scholar]
- 46. Haberle V, Forrest AR, Hayashizaki Y, Carninci P, Lenhard B. 2015. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 43:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohmiya H, et al. 2014. RECLU: a pipeline to discover reproducible transcriptional start sites and their alternative regulation using capped analysis of gene expression (CAGE). BMC Genomics. 15:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the Supplementary material.