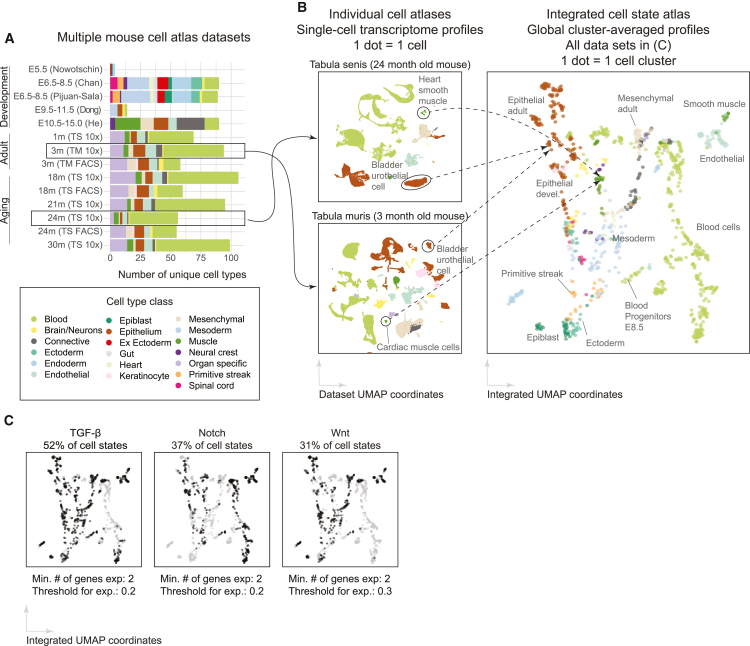
Figure 2.
Integration of scRNA-seq atlas data reveals widespread expression of signaling pathway components
(A) We integrated seven published developmental and adult scRNA-seq datasets spanning 14 different stages in the mouse lifespan from embryonic development to old age. These datasets differ in their representation of organs and cell type classes (colors). The name of the dataset indicates the developmental stage and the first author’s name. For the Tabula Muris and Tabula Muris Senis datasets, we used the abbreviations TM and TS, accordingly.
(B) To generate an integrated cell state atlas, we first independently clustered each scRNA-seq dataset, treating distinct time points in each dataset separately (UMAPs, left; STAR Methods). We then averaged expression over all cells in each cluster to yield a “cell state” profile for that cluster, and we represented each cluster by a single dot in an integrated cell state atlas dataset (UMAP, right). Colors are consistent with the legend in (A).
(C) Components of core signaling pathways are broadly expressed. Black or gray dots show clusters whose pathway components are expressed above or below threshold, respectively.