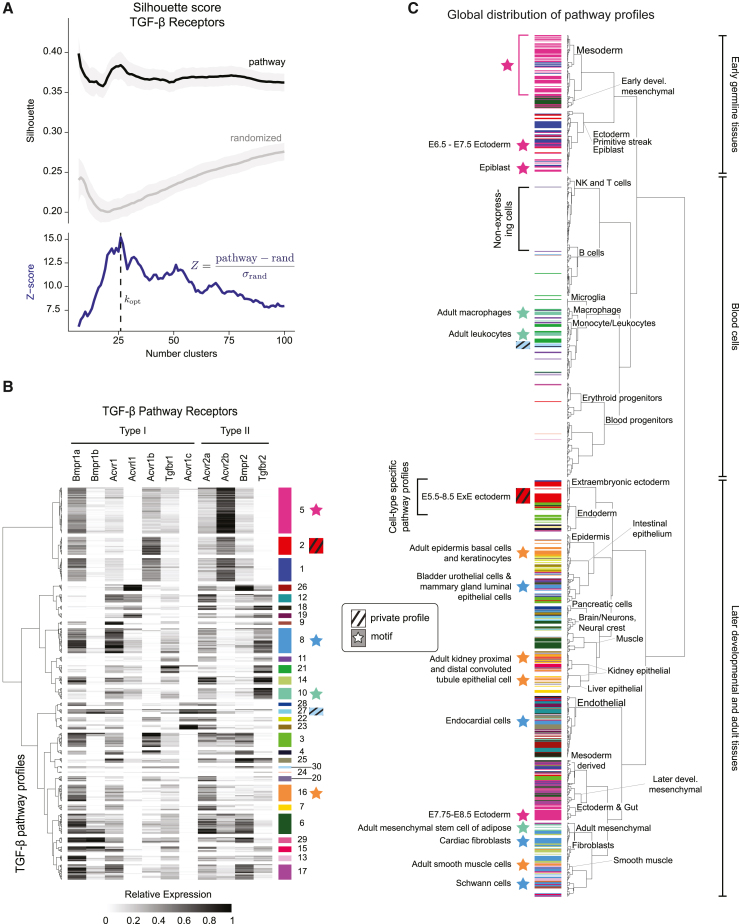
Figure 3.
TGF-β receptors exhibit recurrent and dispersed pathway expression profiles
(A) The silhouette score identifies the approximate number of unique TGF-β receptor expression profiles. We computed the silhouette score across expression values of the pathway genes (black), as well as for 100 random gene sets (gray) where pathway gene expression was independently scrambled for each gene. We then computed the Z score (blue), defined as the silhouette score for pathway genes normalized to the silhouette score for randomized gene sets. We defined the optimal number of receptor profiles as the number of clusters that produced the peak Z score value, in this case, approximately 30 TGF-β receptor expression profiles (dashed line).
(B) Heatmap indicates gene expression of TGF-β receptor components in the 622 cell types expressing the pathway (Figure 2C). The identified ∼30 TGF-β receptor expression profiles are indicated as color-labeled groups of rows. Colored stars indicate examples of dispersed profiles highlighted on the global cell fate dendrogram in (C). Colored shaded boxes indicate private profiles, also shown in (C). Dendrogram at left represents similarity among different profiles. Each gene is standardized to a range of 0–1 across all cell types (grayscale) as described in the STAR Methods.
(C) Distribution of TGF-β receptor expression profiles across cell types. The global cell type dendrogram was computed using a cosine distance metric applied to the integrated transcriptome dataset in a 20-component principal-component analysis (PCA) space constructed from 4,000 highly variable genes (HVGs). Stars indicate featured TGF-β profiles that are broadly dispersed across cell types, while colored shaded boxes indicate examples of private profiles. Cell types that do not express TGF-β receptors have no color (white). Colors match those in (B). Note that blood cell types are relatively lacking in expression of TGF-β receptors.