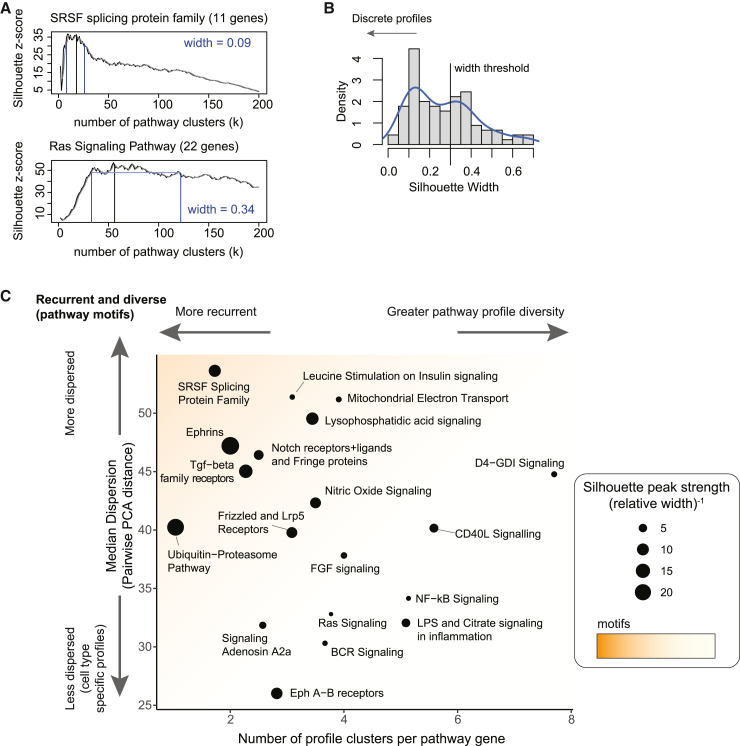
Figure 5.
Expression motifs occur in multiple pathways
(A) The silhouette score calculated for different numbers of clusters (k) normalized as a Z score compared to randomized profiles. The peak width is defined as the number of k values with a Z score within 10% of the maximum Z score (blue lines), relative to the total number of k values evaluated (200). Pathways with a well-defined peak (SRSF, top) display a well-defined number of profiles around the peak. On the other hand, a broad range of k with high silhouette scores indicates higher order structure as for increasing number of clusters (Ras signaling pathway, bottom).
(B) The distribution of width scores for selected pathways (from PathBank database, Table S3).
(C) Dispersion and recurrence metrics for multiple pathways with well-defined peaks (relative width < 0.35). Based on the silhouette Z score, we identified the optimal number of clusters and computed the dispersion for different pathways (y axis). The optimal value of k is normalized by the number of genes in the pathway (x axis). We defined the silhouette peak strength as the inverse of the peak width (dot size). Pathways including motifs appear in the upper-left corner: they display a few discrete profiles that are expressed across multiple cell types.