Figure 7.
Developmental transitions of pathway profiles and summary
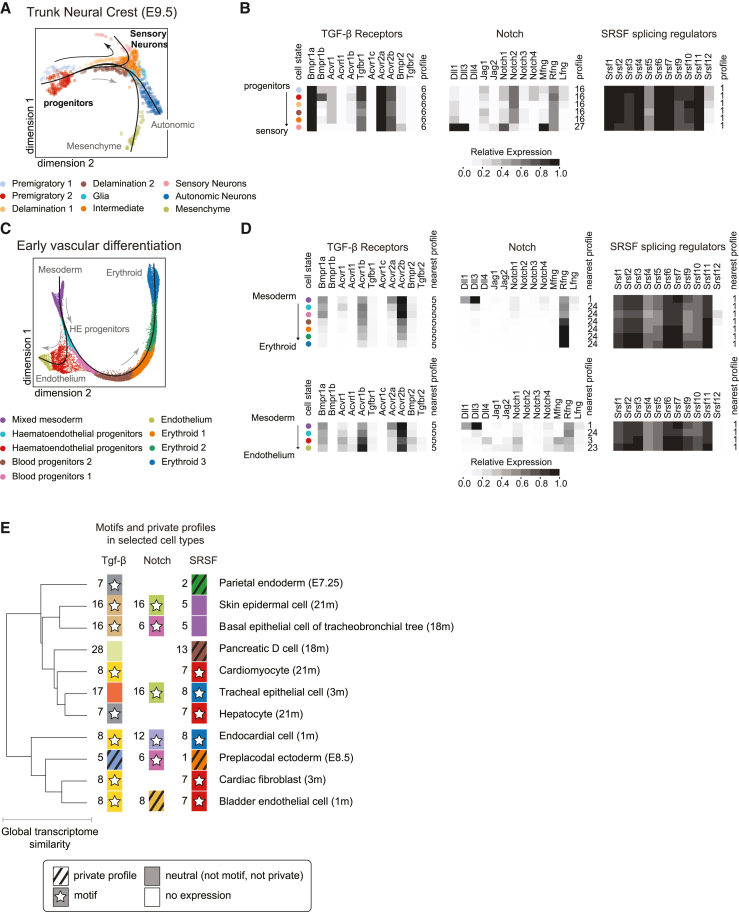
(A) Pseudotime trajectory analysis of the trunk neural crest60 captures delamination of progenitors into three distinct cell fates in a ForceAtlas projection: sensory neurons, autonomic neurons, and the mesenchyme. Here, we follow the sensory neuron trajectory (black arrow).
(B) Developmental pathways expression dynamics in neural crest differentiation. For each pathway, corresponding mean expression profiles are shown in grayscale for each of the cell states indicated in (A), as indicated by the colored dots. Profile numbers indicate the closest match (nearest neighbor) to one of the reference pathway profiles in Figure 3B for TGF-β and Data S2 for Notch and SRSF.
(C) ForceAtlas projection and pseudotime reconstruction of early vascular differentiation.29 Mesodermal progenitors differentiate into endothelial and erythroid cell fates (gray arrows).
(D) Dynamics of three core pathways for each of the two trajectories in (C): erythroid differentiation (upper row of heat maps) and endothelial differentiation (lower row). Colored dots indicate cell states in (C). Profile numbers indicate closest matches in reference profiles (Figure 3B for TGF-β and Data S2 for Notch and SRSF).
(E) Mosaic view of profile usage (schematic). Cell states can express each of their pathways, using any of the distinct available profiles (indicated schematically by profile ticks). In this way, cell states can be thought of, in part, as mosaics built from combinations of available pathway profiles.