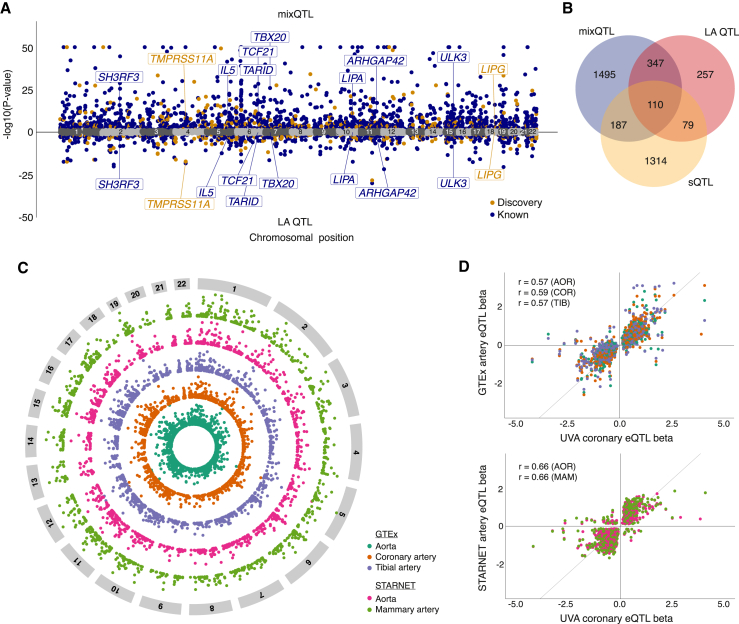
Figure 2.
Overview of eQTL analysis and generalization to published arterial eQTLs
(A) Miami plot of lead eQTLs for mixQTL (top) and local ancestry (LA) adjusted (bottom). Navy blue and orange dots represent reported and discovery eGenes Benjamini-Hochberg adjusted p-value (pBH) < 0.05, respectively; gray dots represent non-significant genes. A subset of top eGenes are labeled for clarity.
(B) Venn diagram showing the overlap of mixQTL and LA-based eGenes and LeafCutter sQTL sGenes.
(C) Circos plot portraying generalized (UVA pBH <0.05) published arterial eGenes from GTEx AOR (blue green), COR (orange), and TIB (purple), and STARNET AOR (pink) and MAM (light green) tissues, with significance increasing toward the outer edge of the circle.
(D) Direction of effect for genes in which the UVA lead eQTL was significant (pBH < 0.05) in any of the aforementioned tissues using the same color scheme for GTEx (top) and STARNET (bottom). Pearson’s r correlation coefficients shown for overlapping significant UVA coronary eQTL detected in GTEx or STARNET eQTL with tissue indicated in parentheses. AOR: aorta; COR: coronary artery; TIB: tibial artery; MAM: mammary artery.