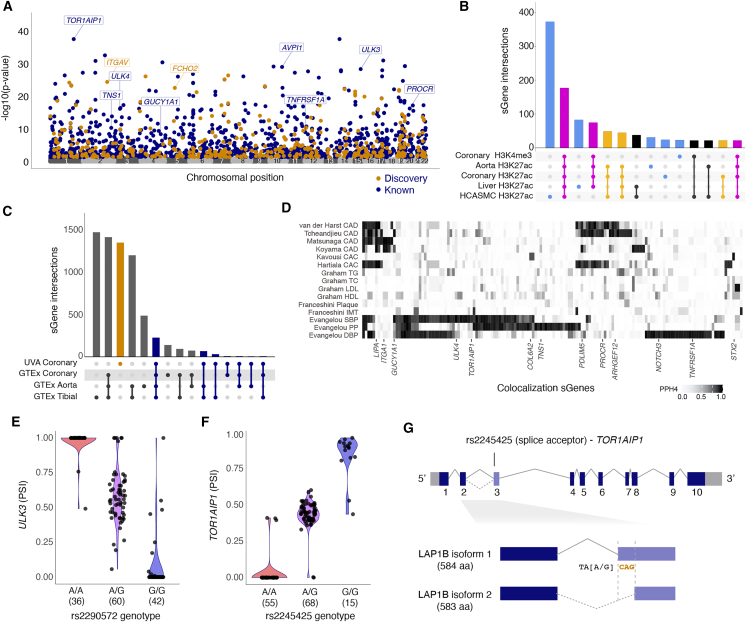
Figure 5.
Coronary artery sQTL overview and characterization
(A) Manhattan plot of lead sQTLs. Navy blue and orange dots represent reported and discovery sGenes (pBH < 0.05), respectively; gray dots represent non-significant genes.
(B) Upset plot of generalization of GTEx arterial sQTLs. Black bars represent GTEx-specific sGenes; orange bars represent generalized sGenes, and the blue bar represents previously unreported sGenes.
(C) Upset plot of histone modifications in aorta, coronary, and liver from one individual in ENCODE as well as HCASMCs. Light blue bars represent lead sQTLs overlapping modifications specific to one cell or tissue type, gold bars for H3K27 acetylation in coronary and any other tissue or cell type, and magenta bars for both coronary artery H3K4 tri-methylation and H3K27 acetylation.
(D) GWAS colocalization to sQTL associations: each row represents the colocalization PPH4 of the study (first author last name) and relevant GWAS trait, with intensity of shading corresponding to a higher posterior probability of a shared association at that locus. Each column represents one protein-coding gene with a PPH4 ≥0.8 in at least one GWAS.
(E and F) Percent spliced in of ULK3 and TOR1AIP1 exons by genotype of their lead sQTLs (rs2290572 and rs2245425, respectively) in our study (left) and GTEx (right).
(G) Schematic of TOR1AIP1 gene and alternatively spliced isoforms, showing location and effect estimates for top splice acceptor variant (rs2245425) identified in our samples. rs2245425-G creates TAGCAG splice acceptor sequence at 3′ end of intron-exon 3 junction. Spliced CAG nucleotides (orange) encoding alanine amino acid distinguish LAP1B isoforms 1 and 2.