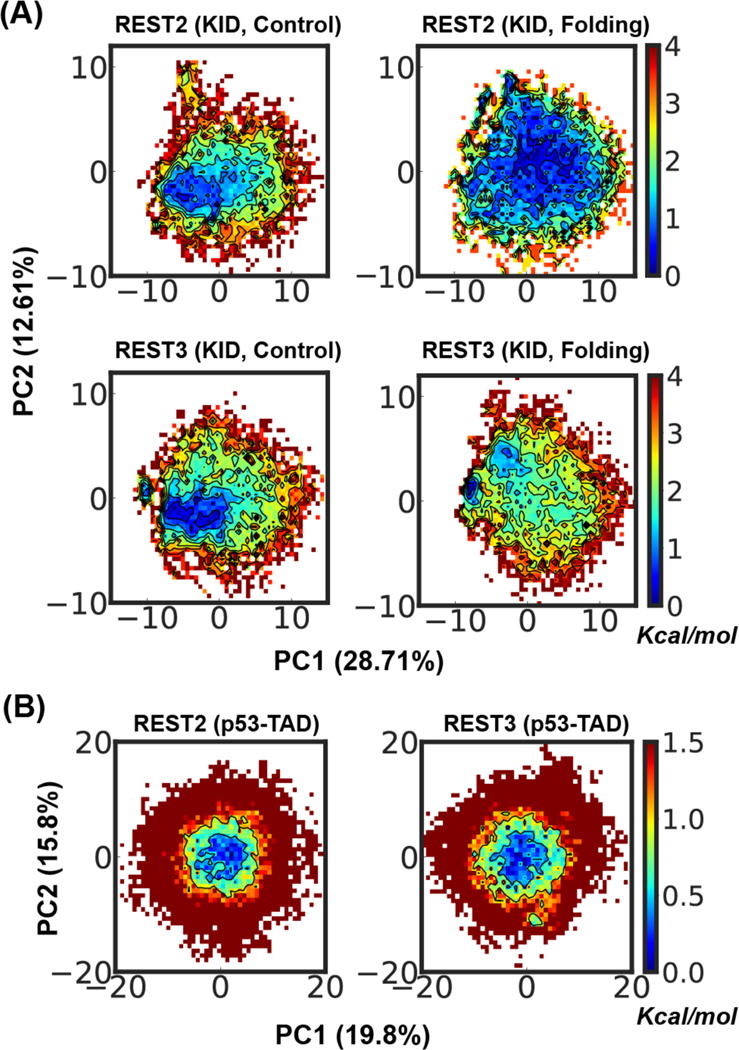
Figure 7.
Distribution of conformational ensembles at 298 K generated by REST2 and REST3 control and folding simulations of KID (A) and folding simulation of p53-NTD (B) in the a99SB-disp force field. The ensembles were projected along the same first two principal components derived from PCA analysis of all conformations sampled from both REST2 and REST3 runs of each individual protein. All probability distributions were first converted into the free energy surface before plotting.