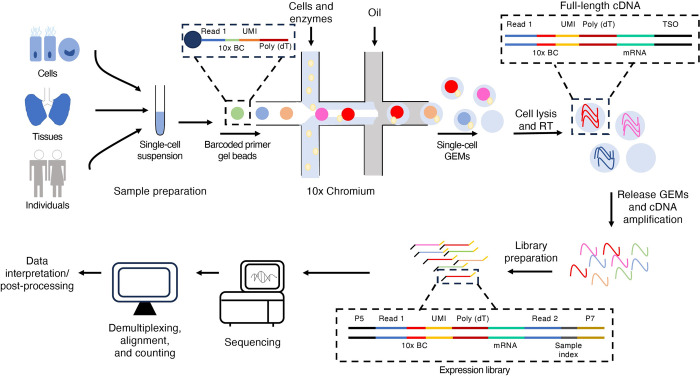
Fig 2. Single-cell sequencing workflow for droplet-based approaches exemplified by the 10x Genomics Chromium platform.
Infected cells from cell culture, tissues/organoids, or infected individuals are dissociated into a single-cell suspension. The cell suspension is loaded onto a microfluidic chip, and cells are partitioned into nanoliter-scale Gel Beads-in-emulsion (GEMs) droplets containing barcoded gel beads and reagents for reverse transcription (RT). Following cell lysis, the beads capture the mRNA molecules. Reverse transcription (RT) by template switching using a template switching oligonucleotide (TS) generates cDNA tagged with a 10x barcode (BC) to identify the cell and a unique molecular identifier (UMI) to label the mRNA transcript. The pooled cDNA is amplified in bulk, fragmented by enzymatic fragmentation, and sequencing adapters (P5 and P7) including a sample index, are added to the fragments by PCR to generate sequencing libraries. The sequencing libraries are sequenced, and the data are analyzed by alignment and demultiplexing, following which the data are interpreted.