ABSTRACT
Background & Aims
Hepatitis delta virus (HDV) infection accelerates the progression of chronic hepatitis B virus (HBV) infection, posing a large economic and health burden to patients. At present, there remains a lack of accurate and portable detection methods for HDV RNA. Here, we aim to establish a convenient, rapid, highly sensitive and specific method to detect HDV RNA using CRISPR–Cas13a technology.
Methods
We established fluorescence (F) and lateral flow strip (L) assays based on CRISPR–Cas13a combined with RT–PCR and RT-RAA for HDV RNA detection, respectively. we conducted a cohort study of 144 patients with HDV-IgG positive to evaluate the CRISPR–Cas13a diagnostic performance for identifying HDV in clinical samples, compared to RT–qPCR and RT-ddPCR.
Results
For synthetic HDV RNA plasmids, the sensitivity of RT–PCR-CRISPR-based fluorescence assays was 1 copy/μL, higher than that of RT–qPCR (10 copies/μL) and RT-ddPCR (10 copies/μL); for HDV RNA-positive samples, the sensitivity of RT-RAA-CRISPR-based fluorescence and lateral flow strip assays was 10 copies/μL, as low as that of RT–qPCR and RT-ddPCR, and the assay took only approximately 85 min. Additionally, the positivity rates of anti-HDV IgG-positive samples detected by the RT–qPCR, RT-ddPCR, RT–PCR-CRISPR fluorescence and RT-RAA-CRISPR lateral flow strip methods were 66.7% (96/144), 76.4% (110/144), 81.9% (118/144), and 72.2% (104/144), respectively.
Conclusions
We developed a highly sensitive and specific, as well as a portable and easy CRISPR-based assay for the detection of HDV RNA, which could be a prospective measure for monitoring the development of HDV infection and evaluating the therapeutic effect.
KEYWORDS: Hepatitis delta virus, CRISPR–Cas13, recombinase-aided amplification, quantitative real-time PCR, droplet digital PCR
Introduction
Hepatitis delta virus (HDV) infection is one of the most serious liver diseases. It is estimated that 15-20 million people are infected with HDV worldwide [1,2]. The HDV genome is a single-stranded negative sense RNA that contains 1672 to 1679 nucleotides, and HDV is a satellite RNA virus that can propagate only in the presence of hepatitis B virus (HBV) [3,4]. HBV coinfection with HDV can dramatically accelerate progression to cirrhosis and the development of hepatocellular carcinoma relative to chronic HBV infection alone. Chronic infection with HDV is considered to be the most severe form of human viral hepatitis infection, significantly increasing the risk of cirrhosis, hepatocellular carcinoma, and acute liver failure, leading to a reduced life expectancy [5,6].
At present, the diagnosis of HDV relies on the total antibody against HDV (anti-HDV), which can be detected by enzyme-linked immunosorbent assay (ELISA) or radioimmunoassay (RIA) [7]. For instance, all individuals infected with HDV are positive for IgG antibodies, which persist after virus clearance; IgM antibodies persist in a large proportion of patients with chronic infection and are considered by some investigators as a surrogate marker for HDV replication [8]. However, the presence of anti-HDV is not always associated with active infection. Within weeks of acute infection, antibodies are still undetectable and do not differentiate between active and resolved infection. Anti-HDV IgM emerges earlier during acute infection but is infrequently detectable in chronic phases and therefore not very useful for confirmation of infection [9]. Currently, the marker that best indicates HDV replication is HDV RNA in the plasma or serum, which is detected in all patients with acute or chronic infection and not detected after spontaneous or treatment-induced viral clearance [3,10].
HDV RNA sequences thus far have been classified into eight known genotypes (HDV-1–8) based on the nucleotide sequence [11]. These genotypes are distributed across different geographical regions. Among them, HDV-1, HDV-2 and HDV-4 are found in Asia [6,11]. Real-time quantitative PCR (RT–qPCR) is the method commonly used to evaluate the HDV-RNA viral load, and several in-house and commercial assays have been developed [10,12,13]. Unfortunately, the results showed high variability due to the heterogeneity of technical tools, including extraction methods, RT–qPCR devices, and internal controls from different laboratories. Recently, we studied the detection of HDV RNA using digital PCR, which allows accurate quantification and has a sensitivity of 1 copy/µL [14]. This provides a new method for the quantitative detection of HDV RNA, but its application cost is high, and it is not conducive to large-scale application. Therefore, it is very important to develop an economical and sensitive detection method for HDV RNA to monitor the occurrence and development of disease and evaluate the therapeutic effect.
The clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein (Cas) system is an acquired immune system for the cleavage of foreign genetic elements from invading viruses and phages and was first identified in bacteria and archaea [15,16]. The CRISPR–Cas system contains several proteins, such as CRISPR/Cas12a, CRISPR/Cas13a and CRISPR/Cas14a, which have been widely applied in nucleic acid analysis for their good specificity and signal amplification ability [17,18]. The CRISPR/Cas13a system recognizes and detects target RNA sequences, and the CRISPR/Cas12a and CRISPR/Cas14a systems recognize and detect target DNA sequences [17,18]. These proteins have trans-cleavage activity and have been utilized to develop diverse nucleic acid detection modalities, including DETECTR, SHELOCK, HOLMES, CARMEN and so on [19–21]. The advantages of the CRISPR–Cas detection system are convenience, cost-effectiveness, high sensitivity and specificity, and it provides a new method for nucleic acid detection.
Here, we describe the development of an efficient, accurate and portable assay for detecting HDV RNA using the CRISPR–Cas13a system combined with RT–PCR and RT-RAA nucleic acid amplification methods. The final results can be displayed by fluorescence readout and a lateral flow strip. This method provides a new measure for monitoring the occurrence and development of HDV infection and evaluating the therapeutic effect.
Materials and methods
Clinical sample collection and ethics approval
Plasma samples were collected from 144 anti-HDV IgG positive patients with HBV-related infection and 10 healthy subjects at Beijing Youan Hospital, Capital Medical University. The HBV viral load and anti-HDV IgG or IgM of these plasma samples were measured by the laboratory department. All plasma samples were collected and stored at – 80°C until processing, and details were anonymized during subsequent laboratory tests for each patient. Ethical approval of the study was given by the Beijing Youan Hospital's human research committee (Ethical code: LL-2020-167-K), and all patients provided informed consent. The procedures followed were in accordance with the ethical standards of the Committee responsible for human experiments and the Declaration of Helsinki of 1975, as amended in 1983.
Plasmid and RNA sample preparations
The target gene fragments of HDV were synthesized and cloned into the pUC57 vector (Biomed Biotechnology, Beijing China). Synthetic DNA targets with a forward primer that contained a T7 promoter sequence were transcribed in vitro to generate synthetic RNA targets. The product was used as the template using the HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, USA) at 37°C overnight.
The transcribed RNA was then treated with RNase-free DNase I (Qiagen, Germany) to remove any remaining DNA according to the manufacturer’s instructions. RNA was purified using RNA Clean XP (Beckman Coulter, USA), quantified by Nanodrop and Qubit and diluted in nuclease-free water to working concentrations. HDV RNA was extracted from 200 µl plasma samples using an automated nucleic acid extraction system (Bioteke, China) according to the manufacturer’s instructions.
According to previous studies, thermal shock was used to disrupt the secondary structure of HDV RNA, that is, 10 µL of HDV RNA at 95°C for 10 min, immediately followed by cooling to −80°C [22,23].
World health organization international standard for HDV
The World Health Organization International Standard (WHO-HDV-IS) for Hepatitis D Virus RNA for Nucleic Acid Amplification Techniques (NAAT)-Based Assays was obtained from the Paul-Ehrlich-Institut (Langen, Germany). The lyophilized plasma standard was resuspended in 0.5 mL molecular biology grade water. RNA was extracted from the WHO-HDV-IS, followed by 10-fold serial dilution (5.75-57500 IU/mL).
Design of RT–PCR, RT-RAA primers and crRNAs
The sequences of the HDV 1-8 genotype were downloaded from the hepatitis delta virus database (https://hdvdb.bio.wzw.tum.de/hdvdb/), and MAFFT version 7 was used for multiple sequence alignment. Three RT–PCR primer sets were designed by Primer3 Plus (https://www.primer3plus.com/index.html). Three RT-RAA primer sets and 4 crRNAs were designed according to previous studies [24]. All primer sets and crRNAs spanned the conserved position in sequences of HDV genotypes and were screened for efficiency in the optimization experiments. All forward primer sets contained a T7 promoter sequence (5’-AATTCTAATACGACTCACTATAGGG-3’), and all sequences were synthesized by Sangon Biotech Co., Ltd. (Shanghai) (Supplementary Tables 1 and 2).
RT–PCR and RT-RAA
RT–PCR was performed using a HiScript II One Step RT–PCR Kit (Vazyme, China) according to the manufacturer’s protocol. The reaction system contained 25 μL of 2× One Step Mix, 2.5 μL of One Step enzyme mix, 400 nM forward and reverse primers, 2 μL of RNA template, and DNase/RNase-free water up to 50 μL. The RT–PCR cycling conditions were as follows: 55°C for 30 min and 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s and then by elongation at 72°C for 5 min. After amplification, 5 μL of the RT–PCR product was analyzed using Cas13a-based detection.
For all the RT-RAA reactions, the RT-RAA kit from Hangzhou ZC Bio-Sci&Tech Co. Ltd. was used according to the instructions. Briefly, 25 μL of Buffer A, 400 nM forward and reverse primers, 5 μL RNA sample and 13.5 μL of RNase-free water were combined into the detection unit tube containing dry reaction powder; 2.5 μL Buffer B was then added to the detection unit's tube cover, the tube cover was closed, and the mixture was well mixed with gentle shaking upside down and rapidly centrifuged at low speed. The reactions were incubated at 39°C for 30 min. After amplification, 5 μL of the RT-RAA product was analyzed using Cas13a-based detection.
Detection assay optimized reaction
The optimized reaction for RT-RAA-CRISPR–Cas13a-based fluorescence detection was performed at the endpoint fluorescence value. In the optimization process, variables were individually controlled, including the RT-RAA reaction time and temperature, CRISPR–Cas13a reaction time and temperature, and concentrations of Cas13a, MgCl2, crRNA and T7 RNA Polymerase.
Fluorescence-based CRISPR–Cas13a detection
The reaction procedure was conducted with the following components: 2 μL ribonucleoside triphosphates (rNTP) (New England Biolabs, USA), 1 μL of RNase inhibitor (New England Biolabs, USA), 45 nM LwCas13a (GenScript, China), 20 mM HEPES buffer solution (ThermoFisher Scientific, USA), 10 mM MgCl2 solution, 1.5 μL crRNA, 0.5 μL of T7 RNA Polymerase (New England Biolabs, Inc.), 2.5 μL RNA reporter (RNAse Alert, Thermo Scientific, Waltham, MA, USA), 5 μL of RT–PCR or RT-RAA amplified product, and 10.75 μL of RNase-free water. The reactions were carried out at 37°C for 60 min using CFX Opus 96 (Bio-Rad, USA) every 2 min.
Lateral flow strip-based CRISPR–Cas13a detection
According to a previous study [25], the reaction procedure was performed with the following components: 4 μL of rNTP (New England Biolabs, USA), 2 μL of RNase inhibitor (New England Biolabs, USA), 45 nM LwCas13a (GenScript, China), 3 μL of crRNA, 1 μL of T7 RNA Polymerase (New England Biolabs, USA), 20 mM HEPES buffer solution (ThermoFisher Scientific, USA), 10 mM MgCl2 solution, 2 nM reporter RNA (5’/6-FAM/UUUUUUUUUUUUUUUUUUUU – Bio/3’), 5 μL of RT–PCR or RT-RAA amplified product, and 26.5 μL of RNase-free water. The reaction tubes were incubated at 37°C for 30 min. After that, the reaction mixture was added to the strip (Hangzhou ZC Bio-Sci&Tech Co. Ltd.), and the results were observed after 3 min. Appearance of the T-band and C-band on the strips indicated negative results, while appearance of only the C-band indicated a positive result. Lateral flow results were assessed by measuring the pixel intensity of the test band using ImageJ.
Evaluation of the CRISPR–Cas13a-based detection assay
In vitro-transcribed HDV RNA and positive samples were serially diluted in water to concentrations of 105, 104, 103, 102, 101, and 100 copies/μL and amplified by RT–PCR or RT-RAA for CRISPR–Cas13a-based detection to evaluate sensitivity. A total of 103 of different viral plasmids (HCV, HEV, HIV) and positive samples were amplified by RT–PCR or RT-RAA for CRISPR–Cas13a-based detection to evaluate specificity.
RT-quantitative real-time PCR (RT–qPCR) for HDV RNA
RT–qPCR was performed using a HiScript II One Step qRT–PCR SYBR Green Kit (Vazyme, China) according to the manufacturer’s protocol. The forward and reverse primers were 5’-GCGAATGGGACCCAGAACTC-3’ and 5’-GTCTCGCGTCCTTCTTTCCT-3’, respectively. The 20 µL qPCR mix comprised 10 µL of 2×One Step SYBR Green Mix, 1 µL One Step SYBR Green Enzyme Mix, 200 nM forward and reverse primers, 2 µL of RNA template, and 6.2 μL of RNase-free water. The RT–qPCR cycling conditions were as follows: RT at 55°C for 3 min, heat activation at 95°C for 30 min and 40 cycles of a denaturing step at 95°C for 10 s followed by annealing and elongation steps at 60°C for 30 s.
RT-droplet digital PCR (RT-ddPCR) to quantify HDV RNA
RT-ddPCR was performed on the TD-1TM Droplet DigitalTM PCR system (TargetingOne, Beijing China) following the manufacturer’s protocol. Thirty microliters of reaction mix containing 7.5 μL of ddPCR Supermix (4×), 400 nM primers (used for RT–qPCR), 200 nM probes and 5 μL of the RNA template was added to the droplet generation chip. Then, 180 μL of droplet generation oil was added, and the chip was used for droplet generation by a Drop Maker. The RT-ddPCR conditions were as follows: 50°C for 5 min and 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s and annealing at 60°C for 1 min and then by cooling at 12°C for 5 min. After that, the amplification products were transferred into a droplet detection chip, which was loaded on a chip reader for fluorescence detection.
Statistical analysis
Data were analyzed by GraphPad Prism 8.0.2 software (GraphPad, Inc., La Jolla, CA, USA). Mean differences in the data were determined by unpaired two-tailed Student’s t tests. The data are represented as the mean ± SEM. Statistical significance was defined as P < 0.05.
Results
Schematic of CRISPR–Cas13a-based detection assay for HDV RNA
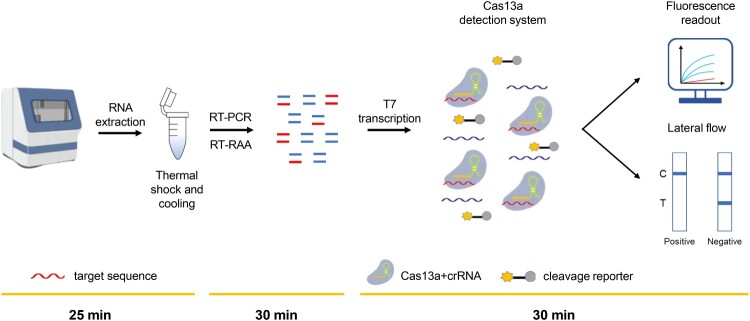
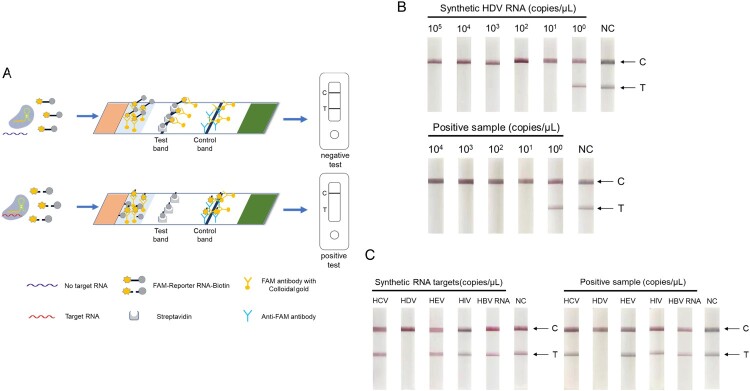
In this study, we developed a novel strategy for the ultrasensitive and fast detection of HDV RNA. As illustrated in Figure 1, total RNA was extracted from the plasma of clinical samples by an automated nucleic acid extraction system, and then was subjected to thermal shock at 95°C, immediately followed by cooling to – 80°C. Subsequently, RT–PCR or RT-RAA amplification was performed using primers targeting the conserved HDV RNA region, which was then converted to RNA by T7 transcription. In the Cas13a detection system, target RNA can be specifically recognized by the Cas13a – crRNA complex to trigger Cas13a activity and to cleave RNA reporters, which are detected by fluorescent instruments or visualized by lateral-flow strips.
Figure 1.
Schematic of the principle of CRISPR–Cas13a-based detection for HDV RNA. Total RNA was extracted from samples using an automated nucleic acid extraction system and subjected to thermal shock and colling (25 min). It is then isothermally amplified to DNA by RT–PCR or RT-RAA (30 min). Then, the DNA is converted to RNA by T7 transcription, and homology binding between the Cas13a-crRNA complex and amplified RNA target triggers the collateral activity of Cas13a, which leads to the cleavage of RNA reporters. The cleaved RNA reporter can be visualized by fluorescence signal detection or captured on a colorimetric lateral flow strip (30 min). C, control line. T, test line.
Specifically, to improve the sensitivity of HDV RNA detection, total RNA was amplified by RT–PCR, and the final fluorescent signal was detected by fluorescent instruments. When total RNA was amplified by RT-RAA, the final fluorescent signal was visualized by a lateral-flow strip, making the detection method faster and more convenient. RNA extraction from plasma samples and thermal shock took 25 min, and RT-RAA amplification and lateral-flow strip detection took 60 min. Thus, the whole process took 85 min, providing a new method for fast and easy detection of HDV RNA in the field.
Design and verification of primers and crRNAs
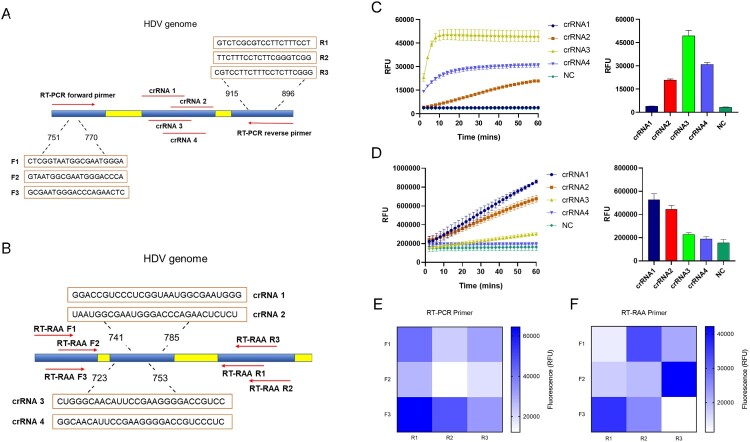
To find the fragment of the relatively conserved region, sequences of HDV genotypes 1-8 were downloaded, and multiple sequence alignment was performed (Supplementary Fig. S1). Three sets of primers and four crRNAs were designed for RT–PCR-CRISPR and RT-RAA-CRISPR assays, respectively (Fig. 2A and B). Screening of four crRNAs for RT–PCR and RT-RAA amplification using RNA templates that were generated through in vitro transcription of synthetic DNA targets was performed with the CRISPR–Cas13a fluorescence assay. As shown in Fig. 2C and D, the kinetics of the fluorescence signal of crRNA3 were the highest for RT–PCR amplification, while crRNA1 demonstrated higher kinetics of the fluorescence signal than the other three crRNAs for RT-RAA amplification. Furthermore, we found that fluorescence signals could be clearly detected in 30 min for two crRNAs.
Figure 2.
Schematic and screening of crRNA and primers for the detection of HDV RNA. A. Location of the crRNA and RT–PCR primer sets for the RT–PCR-CRISPR assay. B. Location of the crRNA and RT-RAA primer sets for the RT-RAA-CRISPR assay. C. Kinetics of fluorescence signal within 60 min and fluorescence values in 30 min of four different crRNAs for RT–PCR-CRISPR reaction. D. Kinetics of the fluorescence signal within 60 min and fluorescence values in 30 min of four different crRNAs for the RT-RAA-CRISPR reaction. E. Heatmaps of 9 different primer pairs for RT–PCR-CRISPR reaction. F. Heatmaps of 9 different primer pairs for the RT-RAA-CRISPR reaction. Forward denoted by “F”, reverse denoted by “R”. The data are the mean ± s.d. for 3 technical replicates. NC, Negative controls.
We then verified the availability of three sets of RT–PCR and RT-RAA primers. The RT–PCR primer pair of F3 and R1 enabled the most efficient target amplification, and the RT-RAA primer pair of F2 and R3 exhibited the best fluorescence efficiency (Fig. 2E and F). Therefore, the crRNA3 and F3R1 primer pair was chosen for the subsequent RT–PCR-CRISPR detection assay, and the crRNA1 and F2R3 primer pair was chosen for the RT-RAA-CRISPR detection assay.
Optimization of the RT-RAA-CRISPR detection assay
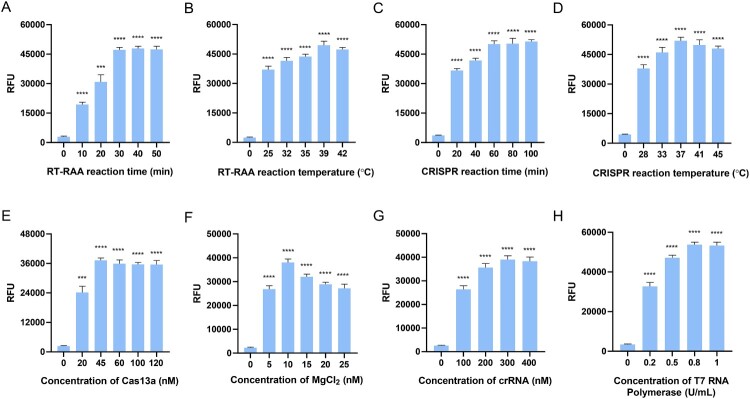
RT-RAA amplification and CRISPR-based detection are enzymatic reactions, and the detection capacity is closely related to parameters such as reaction time, temperature, concentration of enzymes and Cas13a protein. To achieve the best detection effect, we optimized and evaluated some conditions related to detection by the fluorescence intensity at the end point. Here, RT-RAA amplification was performed by incubating from 0 to 50 min at 25°C, 32°C, 35°C, 39°C, and 42°C, and the fluorescence signal value reached a maximum at 30 min and 39°C (Fig. 3A and B). For the CRISPR–Cas13a reaction time and temperature, the fluorescence intensity peaked at 60 min and 37°C. Thus, 60 min and 37°C were selected as the optimal amplification conditions in the following experiments (Fig. 3C and D). Additionally, the concentrations of the Cas13a protein, MgCl2, crRNA, and T7 RNA polymerase were also optimized by detecting the fluorescence intensity. The results show that the fluorescence signal value peaked with a Cas13a concentration of 45 nM. Therefore, 45 nM Cas13a was chosen as the optimal concentration (Fig. 3E), and 10 nM MgCl2, 300 nM crRNA and 0.8 U/mL T7 RNA polymerase were selected as the optimal reaction conditions for subsequent experiments (Fig. 3F-H).
Figure 3.
Optimization of the reaction conditions for the RT-RAA-CRISPR assay. A. Optimizing the reaction time of RT-RAA from 10, 20, 30, 40, 50 min. B. Optimizing the reaction temperature of RT-RAA from 25°C, 32°C, 35°C, 39°C, 42°C. C. Optimizing the reaction time of CRISPR–Cas13a from 20, 40, 80, 60, and 100 min. D. Optimizing the reaction temperature of CRISPR–Cas13a from 28°C, 33°C, 37°C, 41°C, and 45°C. E. Optimizing the concentration of Cas13a from 20, 45, 60, 100, and 120 nM. F. Optimizing the concentration of MgCl2 from 5, 10, 15, 20, 25 nM. G. Optimizing the concentration of crRNA from 100, 200, 300, 400 nM. H. Optimizing the concentration of T7 RNA polymerase from 0.2, 0.5, 0.8, 1 U/mL. The data are the mean ± s.d. for 3 technical replicates. ***: p < 0.001, ****: p < 0.0001.
Evaluation of the CRISPR–Cas13a-based assay using synthetic HDV targets
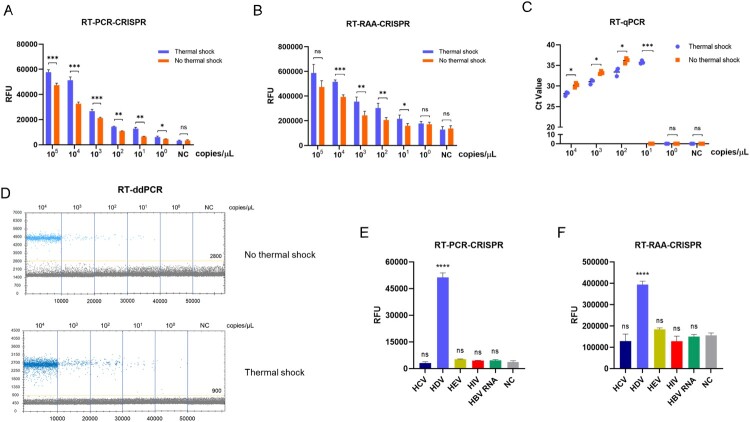
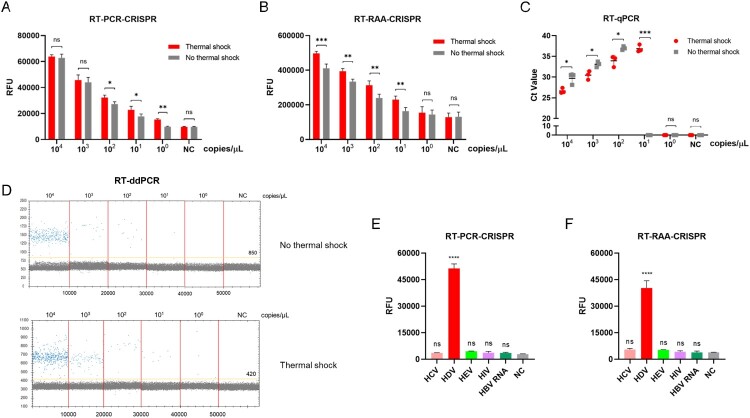
We next evaluated the sensitivity and specificity of CRISPR–Cas13a-based detection with the most promising primer pairs, crRNA and optimal reaction conditions. Synthetic RNA targets were serially diluted for RT-RPA and subsequent detection with and without thermal shock. As illustrated in Fig. 4, when thermal shock and cooling was performed, the limit of detection (LoD) of the RT–PCR-CRISPR method for synthetic HDV RNA was 1 copy/μL with thermal shock, which was the same as RT-ddPCR (y = 1.284x – 0.709, R² = 0.9204) and higher than that of the RT–qPCR (y = −0.3938x + 15.132, R² = 0.997) detection approaches (10 copies/μL) (Fig. 4A, C and D). However, the LoD of the RT–PCR-CRISPR method for synthetic HDV RNA was only 10 copies/μL without thermal shock. Additionally, the fluorescence signal detected showed that the LOD of RT-RAA-CRISPR was the same as RT–qPCR method for synthetic HDV RNA with 10 copies/μL with thermal-shock treatment, while the LOD of RT-RAA-CRISPR was 100 copies/μL without thermal shock (Fig. 4B, C and D).
Figure 4.
Evaluation of RT–PCR-CRISPR and RT-RAA-CRISPR assays using synthetic HDV targets. A. Fluorescence values of RT–PCR-CRISPR for synthetic HDV target serial dilutions with and without thermal shock. B. Fluorescence values of RT-RAA-CRISPR for synthetic HDV target serial dilutions with and without thermal shock. C. Ct values of RT–qPCR for synthetic HDV target serial dilutions with and without thermal shock. D. Scatter plot of ddPCR for synthetic HDV target serial dilutions with and without thermal shock. E. Fluorescence values of RT–PCR-CRISPR for different synthetic targets (HCV, HEV, HIV, HBV RNA). F. Fluorescence values of RT-RAA-CRISPR for different synthetic targets (HCV, HEV, HIV, HBV RNA). The data are the mean ± s.d. for 3 technical replicates. NC, Negative controls. *: p < 0.05, **: p < 0.01, ***: p < 0.001.
To estimate the specificity, hepatitis C virus (HCV), hepatitis E virus (HEV), human immunodeficiency virus (HIV) and HBV RNA gene fragments were synthetized, and the end-point fluorescence intensity demonstrated that RT–PCR-CRISPR or RT-RAA-CRISPR was specific to HDV RNA with no cross-reactivity (Fig. 4E, F).
Evaluation of the CRISPR–Cas13a-based assay using an HDV-positive sample
To further verify the sensitivity and specificity of CRISPR–Cas13a-based detection, we extracted RNA from clinical positive samples. Using serial dilutions of HDV RNA with and without thermal shock, we discovered that RT–PCR-CRISPR achieved a sensitivity as high as 1 copy/μL, which was was the same as ddPCR and higher than that of RT–qPCR 10 copies/μL when thermal shock was performed, and the sensitivity of RT–PCR-CRISPR was only 10 copies/μL without thermal shock (Fig. 5A, C and D). Moreover, the fluorescence signal of RT-RAA-CRISPR was the same as that of the RT–qPCR assays, with a value of 10 copies/μL with thermal shock, while the LOD of RT-RAA-CRISPR was 100 copies/μL without thermal shock (Fig. 5B, C and D).
Figure 5.
Evaluation of RT–PCR-CRISPR and RT-RAA-CRISPR assays using HDV-positive samples. A. Fluorescence values of RT–PCR-CRISPR for HDV-positive sample serial dilutions. B. Fluorescence values of RT-RAA-CRISPR for HDV-positive sample dilutions. C. Ct values of RT–qPCR for HDV-positive sample serial dilutions. D. Scatter plot of ddPCR for HDV-positive sample serial dilutions. E. Fluorescence values of RT–PCR-CRISPR for different positive samples (HCV, HEV, HIV, HBV RNA). F. Fluorescence values of RT-RAA-CRISPR for different positive samples (HCV, HEV, HIV, HBV RNA). The data are the mean ± s.d. for 3 technical replicates. NC, Negative controls. *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Clinical positive samples of HCV, HEV, HIV and HBV RNA were extracted to explore specificity. The results show the high specificity of this method (Fig. 5E, F). Thus, the CRISPR–Cas13a-based assay also has high sensitivity and specificity for clinically positive samples.
Evaluation of the CRISPR–Cas13a-based assay using WHO-HDV-IS
We next used WHO-HDV-IS to measure the sensitivity of RT–qPCR and RT–PCR-CRISPR. As shown in Supplementary Fig. S2, after thermal shock and cooling, the sensitivity of the RT–PCR-CRISPR method to detect WHO-HDV-IS was 5.75 IU/mL, which was higher than that of the RT–qPCR method (57.5 IU/mL). Therefore, our established RT–PCR-CRISPR assay has the same high sensitivity for the detection of WHO-HDV-IS.
CRISPR–Cas13a-based lateral-flow detection
To explore rapid and portable testing of HDV RNA, we combined RT-RAA-CRISPR detection with colloidal gold-based lateral flow strips. According to previous research, the schematic diagram is shown in Figure 6A. The RNA reporter is functionalized with biotin and FAM-reporter, allowing capture on a lateral-flow strip with streptavidin and detection with FAM antibody conjugated to colloidal gold. When HDV RNA targets were recognized by crRNA, the collateral activity of the Cas13a protein cleaved the RNA reporter. FAM antibody with colloidal gold cannot bind to the test line for “positive”, binding instead on the “negative” line. We then evaluated the efficiency of the RT-RAA-CRISPR-lateral flow strip. The results suggested that the LoD of this method was 10 copies/μL for both synthetic targets and clinical positive samples (Fig. 6B). Additionally, the lateral flow strip did not detect any other virus targets and showed high specificity (Fig. 6C).
Figure 6.
RT-RAA-CRISPR-lateral flow strip assay for HDV RNA. A. Illustration of the Cas13a-crRNA combined lateral flow strip assay. B. Lateral flow strip detection of synthetic HDV RNA and HDV-positive sample serial dilutions using RT-RAA-CRISPR after 30 min. C. Lateral flow strip detection of synthetic RNA and positive samples (HCV, HEV, HIV, HBV RNA) using RT-RAA-CRISPR after 30 min. The data are the mean ± s.d. for 3 technical replicates. C, control line. T, test line.
Performance of CRISPR–Cas13a-based assay in clinical samples
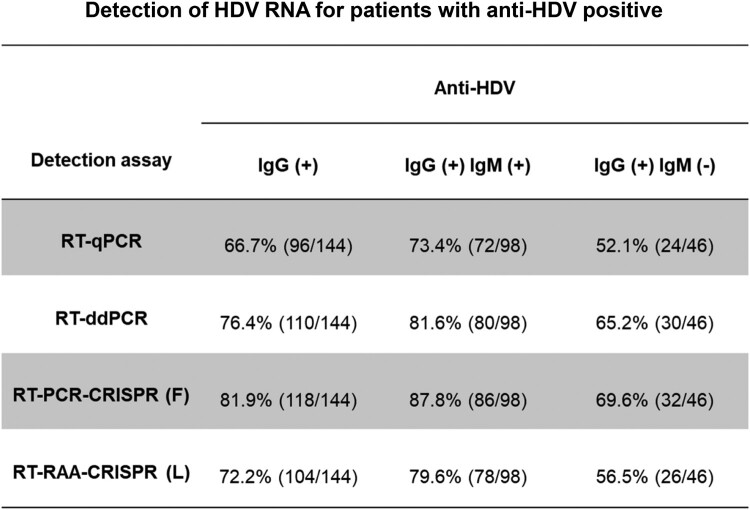
We then tested HDV RNA in plasma from 144 anti-HDV-positive clinical patients with RT–qPCR, RT-ddPCR, RT–PCR-CRISPR-fluorescence (F) and RT-RAA-CRISPR-lateral flow strip (L). Anti-HDV in all clinical samples was detected by the Clinical Laboratory Center of Beijing YouAn Hospital, Capital Medical University (Supplementary Table 3). Among 144 anti-HDV-positive patients, 96 HDV RNA samples were detected by RT–qPCR (positive rate: 66.7%, 96/144), 110 samples were detected by RT-ddPCR (positive rate: 76.4%, 110/144), 118 samples were detected by RT–PCR-CRISPR (F) (positive rate: 81.9%, 118/144), and 104 samples were detected by RT-RAA-CRISPR (L) (positive rate: 72.2%, 104/144). Among 98 anti-HDV IgG – and IgM-positive samples, the positive rate was 73.4% (72/98) by RT–qPCR, 81.6% (80/98) by RT-ddPCR, 87.8% (86/98) by RT–PCR-CRISPR (F), and 79.6% (78/98) by RT-RAA-CRISPR (L). (Fig. 7). The results suggested that the CRISPR-based assay has a high positive detection rate for HDV RNA in clinical samples, while the RT-RAA-CRISPR (L) assay offers faster detection times and easier operation, which is more conducive to testing in the field.
Figure 7.
Detection of HDV RNA for patients with anti-HDV positive. Among the 144 HDV antibody positive samples, 98 samples were positive for both IgG and IgM. RT-qPCR, RT-ddPCR, RT-PCR-CRISPR (F) and RT-RAA-CRISPR (L) methods were used for detection.
Discussion
In this study, we developed a CRISPR–Cas13a-based detection method for HDV RNA combined with RT–PCR or RT-RAA amplification. Total RNA from plasma samples was extracted by an automated nucleic acid extraction system and subjected to thermal shock, which took 25 min; RT–PCR or RT-RAA assay was then performed to amplify HDV RNA targeted fragments, which took 30 min; the target RNA was then detected using the CRISPR–Cas13a system, and the results were determined by fluorescent instruments or lateral-flow strips, which took 30 min. The detection of synthetic plasmids and clinical positive samples revealed that the RT–PCR-CRISPR fluorescence method has high sensitivity and specificity, while the RT-RAA-CRISPR lateral-flow strip assay can provide accurate and portable detection outcomes within 85 min. The difference between RT-PCR-CRISPR and RT-RAA-CRISPR is the amplification method, one is by RT-PCR amplification, the other is by RT-RAA amplification. RT-PCR is a technique that cDNA is used as a template for denaturation, annealing and extension under the action of DNA polymerase to amplify the target fragment. RT-RAA is an at-constant temperature rapid nucleic acid amplification technique that utilizes recombinases, single-stranded DNA binding proteins, DNA polymerases and reverse transcriptase obtained from bacteria to amplify the target fragments at a constant temperature of 30-42°C. The similarity is that both methods use CRISPR technology for detection. RT-PCR-CRISPR technology has better sensitivity and detection rate than RT-RAA-CRISPR technology, but RT-RAA-CRISPR technology is easier to operate and saves time. Therefore, RT-PCR-CRISPR technology is more suitable for clinical testing, while RT-RAA-CRISPR technology is more suitable for on-site testing and screening.
Currently, the main method of HDV screening is by detecting total HDV antibody (anti-HDV), including by ELISA or RIA, but it may not be detected in the initial weeks of acute infection [26]. Confirmation of HDV infection is based on detection of HDV RNA by quantitative polymerase chain reaction (PCR). PCR plays a critical role in the diagnosis and therapeutic monitoring of HDV-infected patients [8,27]. A previous study reported that a one-step TaqMan quantitative reverse transcription-polymerase chain reaction (qRT–PCR) assay was developed for the detection of HDV RNA. The limit of detection of this assay was 7.5×102 copies/ml, and a 31.1% HDV RNA-positive rate was detected in anti-HDV-positive specimens [28]. However, on the basis of the nucleotide and amino acid sequences of the hepatitis delta antigen coding region, HDV RNA sequences are classified into eight genotypes (HDV 1-8), which are placed in different geographical regions [29]. A significant number of studies have not detected all HDV genotypes. In this study, we downloaded the sequences of eight HDV genotypes from the HDV database and performed sequence alignment analysis to find a relatively conserved region in which the primers and crRNAs were designed. Thus, our established CRISPR–Cas13a-based assay improves the sensitivity and specificity of HDV RNA detection.
Although many studies have utilized RT–qPCR methods for detection and quantification, there is a lack of standardized PCR techniques for HDV RNA used in different laboratories [26]. In 2013, the 1st WHO-HDV-IS for HDV RNA was developed to improve the standard uniformity of detection methods. Previous studies have reported the detection of WHO-HDV-IS using RT–qPCR, sequencing and droplet digital PCR assay, which has been validated to allow result comparison between the available detection techniques [10,14,30]. Our study revealed that the sensitivity of the RT–PCR-CRISPR method for detecting WHO-HDV-IS was 5.75 IU/mL, which was higher than that of the RT–qPCR method (57.5 IU/mL). Furthermore, effective quantification of HDV RNA may be compromised by the presence of 70% internal base pairing in the HDV genome. Researchers have used thermal shock to disrupt the structure of the HDV RNA genome and improve the efficiency of detection [22,23,31]. In our studies, synthetic HDV targets and HDV-positive samples were subjected to thermal shock. We demonstrated that the sensitivity of RT–PCR-CRISPR, RT-RAA-CRISPR and RT–qPCR assays with thermal shock was higher than that without thermal shock.
As an efficient detection tool, the CRISPR–Cas13a system has been used for the detection of a variety of disease pathogens, such as some influenza viruses, dengue viruses, and SARS-CoV-2, which have emerged in recent years [32–34]. In a previous study, we demonstrated that the sensitivity of the CRISPR–Cas13a assay was 1 copy/μL for HBV covalently closed circular DNA (cccDNA), and the positive coincidence rate in clinical liver tissue samples was higher than that obtained by other methods; we formerly established an efficient CRISPR–Cas13a assay for HBV DNA with low-level viremia (LLV) [24, 35]. In this study, we developed RT–PCR-CRISPR and RT-RAA-CRISPR assays, both of which allow highly sensitive and specific detection of HDV RNA in less than 85 min. More importantly, the RT-RAA-CRISPR assay combined with a lateral flow strip is convenient and can be quickly performed, requiring only a small thermostat for a high-performance assay. We tested the plasma of 72 anti-HDV-positive patients using RT–qPCR, RT-ddPCR, RT–PCR-CRISPR (F) and RT-RAA-CRISPR (L) assays. Our data revealed that compared with the RT–qPCR assay, RT–PCR-CRISPR (F) and RT-RAA-CRISPR (L) had higher sensitivity and specificity than ddPCR; RT–PCR-CRISPR (F) had higher sensitivity and lower specificity than RT-RAA-CRISPR (L), which may be due to the better fluorescence recognition capability of the fluorescence detector. Anti HDV antibodies are the preferred screening method for detecting HDV infection. However, HDV-IgG positive may also indicate previous infection, the detection result of HDV-IgG may be false positive. For the detection of such clinical samples, RT-qPCR, RT-ddPCR and RT-PCR-CRISPR (F) methods have low accuracy, while RT-RAA-CRISPR (L) method can reduce false positives and improve accuracy due to the flow test strip technology. However, whether it is infected with HDV should be analyzed in combination with the clinical information of the patient.
The cost analysis shows that the automatic nucleic acid extraction equipment is thousands of dollars, and future research can develop nucleic acid lysates, which will greatly reduce the cost. The constant temperature heating device is $ 10, and lateral-flow strip is less than one dollar each. The reagent cost is $ 3 per test. The whole process is convenient and quick to operate, and does not require professionals, which is conducive to testing at different healthcare settings.
This study had some limitations. For total RNA extraction from clinical plasma samples, we used an automated nucleic acid extraction instrument, which was less time consuming but was more expensive, larger, and not suitable for handling or on-site testing. There is a need to continue to explore more convenient and rapid methods for nucleic acid extraction. Additionally, the CRISPR–Cas13a-based assay does not allow precise quantification of HDV RNA, and further studies are required to resolve this concern.
In summary, we developed novel RT–PCR-CRISPR (F) and RT-RAA-CRISPR (L) assays for the detection of HDV RNA, which is not only highly sensitive and specific but also less time-consuming and more conveniently performed. This study provides a powerful tool for monitoring the emergence and progression of disease and for evaluating the effectiveness of treatment.
Supplementary Material
Funding Statement
This study was supported by the National Natural Science Foundation of China (grand number: 82002243); Key Projects of the Beijing Municipal Education Commission's Science and Technology Plan (grand number: KZ202010025035); Special key research project of capital health development scientific research (grand number: SF2020-1-1151, SF2022-1-2182); Beijing Talents foundation, (grand number: 2018000021469G289); Beijing Hospitals Authority Youth Programme (grand number: QML20201702); The Beijing Municipal Institute of Public Medical Research Development and Reform Pilot Project (grand number: JYY2021-10). Talent Cultivation plan of “Climbing the peak” of Beijing Municipal Hospital Administration (grand number: DFL20221503). High-level public health technical personnel construction Project (Subject leaders-02-13). Research and cultivation foundation of capital medical university (grand number: PYZ22130). Capital Clinical Characteristic Diagnosis and Treatment Technology Research and Transformation Application (grand number: Z211100002921051). Beijing You'an Hospital, Capital Medical University-Young and middle-aged talents incubation project (YNKTLC2021002).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
FR, HL and ZPD designed the paper. YT, ZHF and XYZ performed and analyzed experiments, and YT wrote the paper. LX, YLC, ZZP, YKM and HBZ analyzed data and reviewed the paper. HL and ZPD reviewed the paper. FR designed, supervised, and analyzed experimental work and paper. All authors have read and approved the submission of the manuscript.
Data availability statement
All data are available in the main text or the supplementary materials.
References
- 1.Lempp FA, Ni Y, Urban S.. Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options. Nat Rev Gastroenterol Hepatol. 2016;13(10):580–589. [DOI] [PubMed] [Google Scholar]
- 2.Miao Z, Zhang S, Ou X, et al. . Estimating the global prevalence, disease progression, and clinical outcome of hepatitis delta virus infection. J Infect Dis. 2020;221(10):1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucifora J, Delphin M.. Current knowledge on hepatitis delta virus replication. Antiviral Res. 2020;179:104812. [DOI] [PubMed] [Google Scholar]
- 4.Sureau C, Negro F.. The hepatitis delta virus: replication and pathogenesis. J Hepatol. 2016;64(1 Suppl):S102–S116. [DOI] [PubMed] [Google Scholar]
- 5.Béguelin C, Moradpour D, Sahli R, et al. . Hepatitis delta-associated mortality in HIV/HBV-coinfected patients. J Hepatol. 2017;66(2):297–303. [DOI] [PubMed] [Google Scholar]
- 6.Rizzetto M, Hamid S, Negro F.. The changing context of hepatitis D. J Hepatol. 2021;74(5):1200–1211. [DOI] [PubMed] [Google Scholar]
- 7.Chen LY, Pang XY, Goyal H, et al. . Hepatitis D: challenges in the estimation of true prevalence and laboratory diagnosis. Gut Pathog. 2021;13(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alfaiate D, Dény P, Durantel D.. Hepatitis delta virus: from biological and medical aspects to current and investigational therapeutic options. Antiviral Res. 2015;122:112–129. [DOI] [PubMed] [Google Scholar]
- 9.Lempp FA, Roggenbach I, Nkongolo S, et al. . A rapid point-of-care test for the Serodiagnosis of hepatitis delta virus infection. Viruses. 2021;13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Gal F, Brichler S, Sahli R, et al. . First international external quality assessment for hepatitis delta virus RNA quantification in plasma. Hepatology. 2016;64(5):1483–1494. [DOI] [PubMed] [Google Scholar]
- 11.Usman Z, Velkov S, Protzer U, et al. . HDVdb: A comprehensive hepatitis D virus database. Viruses. 2020;12(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferns RB, Nastouli E, Garson JA.. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J Virol Methods. 2012;179(1):189–194. [DOI] [PubMed] [Google Scholar]
- 13.Scholtes C, Icard V, Amiri M, et al. . Standardized one-step real-time reverse transcription-PCR assay for universal detection and quantification of hepatitis delta virus from clinical samples in the presence of a heterologous internal-control RNA. J Clin Microbiol. 2012;50(6):2126–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L, Zhang X, Cao Y, et al. . Digital droplet PCR for detection and quantitation of hepatitis delta virus. Clin Transl Gastroenterol. 2022;13(7):e509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amitai G, Sorek R.. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14(2):67–76. [DOI] [PubMed] [Google Scholar]
- 16.Sander JD, Joung JK.. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski MM, Abudayyeh OO, Gootenberg JS, et al. . CRISPR-based diagnostics. Nat Biomed Eng. 2021;5(7):643–656. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Shang X, Huang X.. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg Microbes Infect. 2020;9(1):1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerman CM, Myhrvold C, Thakku SG, et al. . Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582(7811):277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JS, Ma E, Harrington LB, et al. . CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gootenberg JS, Abudayyeh OO, Lee JW, et al. . Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homs M, Giersch K, Blasi M, et al. . Relevance of a full-length genomic RNA standard and a thermal-shock step for optimal hepatitis delta virus quantification. J Clin Microbiol. 2014;52(9):3334–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lempp FA, Schlund F, Rieble L, et al. . Recapitulation of HDV infection in a fully permissive hepatoma cell line allows efficient drug evaluation. Nat Commun. 2019;10(1):2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Y, Fan Z, Xu L, et al. . CRISPR/Cas13a-assisted rapid and portable HBV DNA detection for low-level viremia patients. Emerg Microbes Infect. 2023;12(1):e2177088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Dong X, Wang Y, et al. . Sensitive and easy-read CRISPR strip for COVID-19 rapid point-of-care testing. Crispr J. 2021;4(3):392–399. [DOI] [PubMed] [Google Scholar]
- 26.Mentha N, Clément S, Negro F, et al. . A review on hepatitis D: from virology to new therapies. J Adv Res. 2019;17:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh C, Heller T, Glenn JS.. Pathogenesis of and new therapies for hepatitis D. Gastroenterology. 2019;156(2):461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kodani M, Martin A, Mixson-Hayden T, et al. . One-step real-time PCR assay for detection and quantitation of hepatitis D virus RNA. J Virol Methods. 2013;193(2):531–535. [DOI] [PubMed] [Google Scholar]
- 29.Gal L, Gault F, Ripault E, et al. . Eighth major clade for hepatitis delta virus. Emerg Infect Dis. 2006;12(9):1447–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pyne MT, Mallory MA, Xie HB, et al. . Sequencing of the hepatitis D virus RNA WHO international standard. J Clin Virol. 2017;90:52–56. [DOI] [PubMed] [Google Scholar]
- 31.Giersch K, Homs M, Volz T, et al. . Both interferon alpha and lambda can reduce all intrahepatic HDV infection markers in HBV/HDV infected humanized mice. Sci Rep. 2017;7(1):3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Xia Q, Wu J, et al. . A sensitive electrochemical method for rapid detection of dengue virus by CRISPR/Cas13a-assisted catalytic hairpin assembly. Anal Chim Acta. 2021;1187:339131. [DOI] [PubMed] [Google Scholar]
- 33.Welch NL, Zhu M, Hua C, et al. . Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants. Nat Med. 2022;28(5):1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H, Bu S, Xu Y, et al. . CRISPR/cas13a combined with hybridization chain reaction for visual detection of influenza A (H1N1) virus. Anal Bioanal Chem. 2022;414(29-30):8437–8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Tian Y, Xu L, et al. . CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection. Hepatol Int. 2022;16(2):306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.