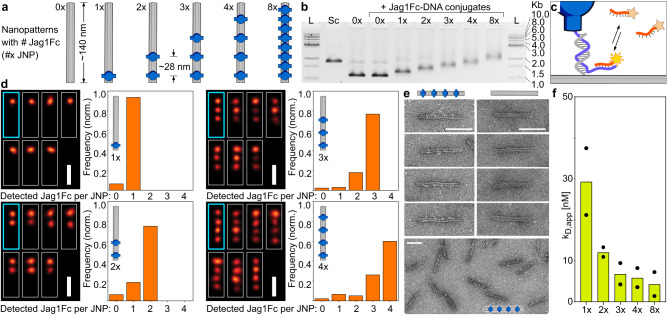
Fig. 2. Characterization of Jag1Fc DNA nano-patterns.
a A rod like DNA origami was used to create 1-8x Jag1Fc nanopatterns (JNPs). b Gel shift assay. 1Kb ladder (L), origami scaffold alone (Sc), empty JNP(0x), a repeat of(0x) but with added Jag1Fc-DNA conjugates (control for non-specific binding), structures loaded with Jag1Fc patterns: 1-8x JNP (2% agarose, EthBr stained). Agarose gel repeated independently for n = 20 c Schematic representation of the DNA origami used for the DNA PAINT experiments. Here, the Jag1-DNA conjugates also contain an extension for DNA-PAINT docking sites. d DNA PAINT on Jag1Fcs on DNA origami Average (cyan, thick) and individual cropped DNA PAINT superresolution images (white, thin) of JNPs (scale bars = 50 nm). Bar graphs show the Jag1 site occupancy distributions of the different nanorods. e Zoom-ins of negative stain TEM of (unpurified) 4x JNPs and empty DNA origami rods (0x JNP) and zoom-out of 4x JNPs (bottom). Scale bars are 100 nm. Micrographs were repeated independently for n = 5 f Receptor immobilized on SPR chip surface and increasing concentrations of 1-8x JNP as analyte. The bar graph shows the mean apparent kD of different Jag1Fc nano-patterns, dots represent two individual repeats.