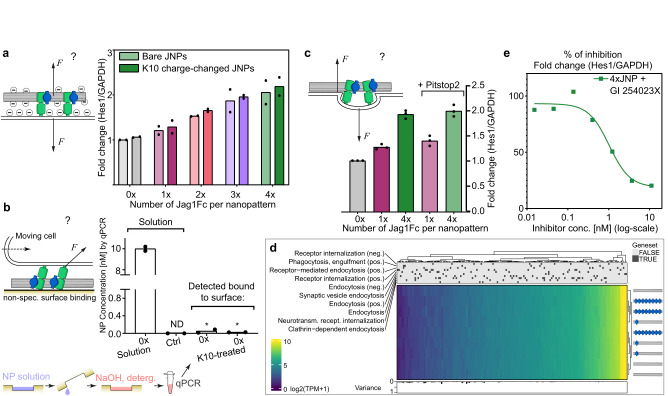
Fig. 4. Multivalency drive Notch activation despite potential force sources and ADAM10 is the driving metalloprotease.
a Electrostatic repulsion between negative DNA origami and negative cell membrane could cause pull. qPCR on lt-NES cells stimulated with normal JNPs, or oligolysine (K10) coated JNPs (charge changed towards neutral). Bar graphs show the mean expression levels and dots represent 2 biological repeats. b Non-specific surface attachment of JNPs could drive force generation in motile cells. qPCR of part of the DNA origami scaffold (M13mp18 ssDNA) to detect concentrations of structures in solution and, retrieved by harsh washing of cell culture surfaces exposed to JNP solution, any remaining non-specifically bound DNA origami. *Denotes measurements below the linear range of the assay. Bar graphs show the mean value of 2 technical repeats shown with dots c Endocytosis could drive force generation. qPCR of HES1 with, or without, Pitstop2 (inhibitor for clathrin-mediated endocytosis) on JNP-stimulated lt-NES cells. Bar graphs show the mean value of 3 technical repeats shown with dots. d RNA sequencing data of gene ontology genesets that are implied in different levels of endocytosis. Data is shown automatically clustered using hierarchical complete-linkage clustering of Euclidean distances. e Increasing concentrations of ADAM inhibitor GI 254023X added on iPS cells and stimulated with 4x JNPs. qPCR of HES1. Square symbols represent the mean value of 3 technical repeats. Source data are provided as a source data file.