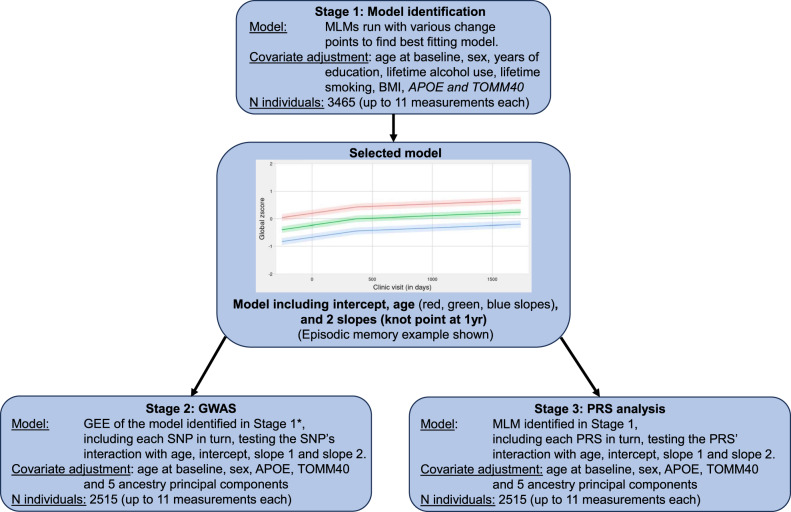
Fig. 1. Schematic of the study design.
Stage 1 involved model identification of the trajectories and determined that models with a knot (change) point at 1 year adequately represented the data. The resulting models were then used in GWAS and PRS analyses (adjusting only for non-heritable confounders and the genetic factors that affected selection into the trial). *Multilevel models were used for initial model exploration and PRS analysis. For the GWAS, the time taken to fit each MLM meant they could not be used. Instead, we used GEE, which treats the correlation between repeated measures as a nuisance parameter and is therefore much faster to fit. MLM multilevel model, GEE generalised estimating equation, GWAS genome-wide association study, SNP single nucleotide polymorphism, PRS polygenic risk score, BMI body mass index.