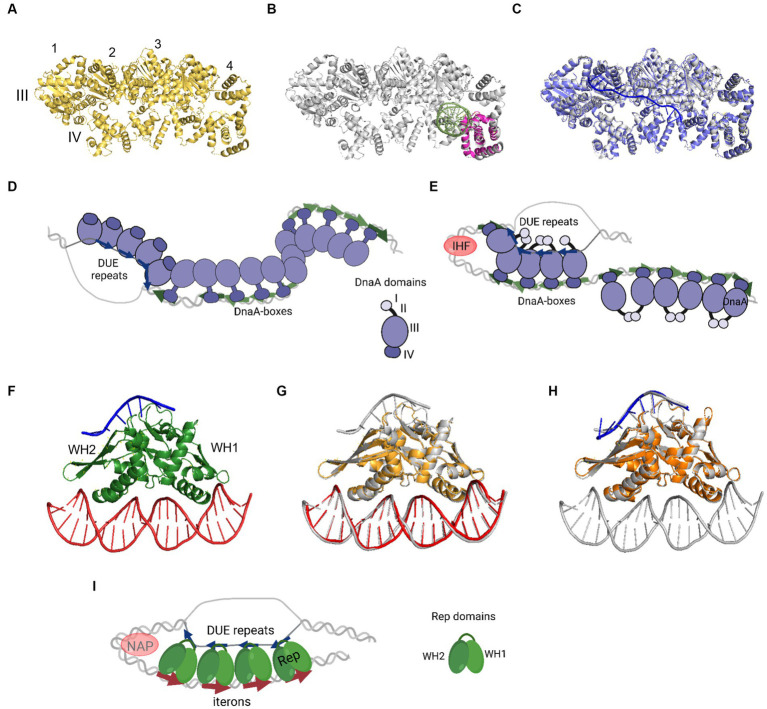
Figure 2.
The nucleoprotein complexes formed by replication initiator proteins bacterial DnaA and plasmid Rep proteins. (A) The oligomeric structure of four protomers of domains III and IV of AaDnaA PDB: 2HCB (yellow). Each protomer is numbered and domains III and IV are indicated. (B) Alignment of PDB: 2HCB structure of AaDnaA (gray) and PDB: 1J1V structure of IV domain of EcDnaA protein (pink) in the complex with dsDNA (light green). (C) Alignment of PDB: 2HCB structure of AaDnaA (gray) and PDB: 3R8F structure of domains III and IV of AaDnaA (light and dark purple) in a complex with ssDNA (blue). (D) The two-state model of bacterial origin opening based on Ekundayo and Bleichert (2019). (E) The loop-back model of bacterial origin opening based on Katayama et al. (2017). (F) The PDB: 8AAN structure of RepE protein (green) in a complex with dsDNA (red) and ssDNA (blue). The WH1 and WH2 domains are indicated. (G) The alignment of PDB: 8AAN tripartite RepE nucleoprotein complex structure (gray) and PDB: 1REP structure of RepE (light orange) in a complex with dsDNA (red). (H) The alignment of PDB: 8AAN tripartite RepE nucleoprotein complex structure (gray) and PDB: 8 AC8 structure of RepE (orange) in a complex with ssDNA (blue). (I) The loop-back model of plasmid origin opening based on Wegrzyn et al. (2023). (D,E,I) DnaA (purple), DnaA-boxes (green arrows), DUE repeats (blue arrows), Rep protein (green), iterons (red arrows), and NAP protein, e.g., IHF (light red) are presented. The alignment of the structures was done with PyMOL software (PyMOL Molecular Graphics System, version 2.4.0, Schrödinger, LLC; http://pymol.org/).