Figure 4.
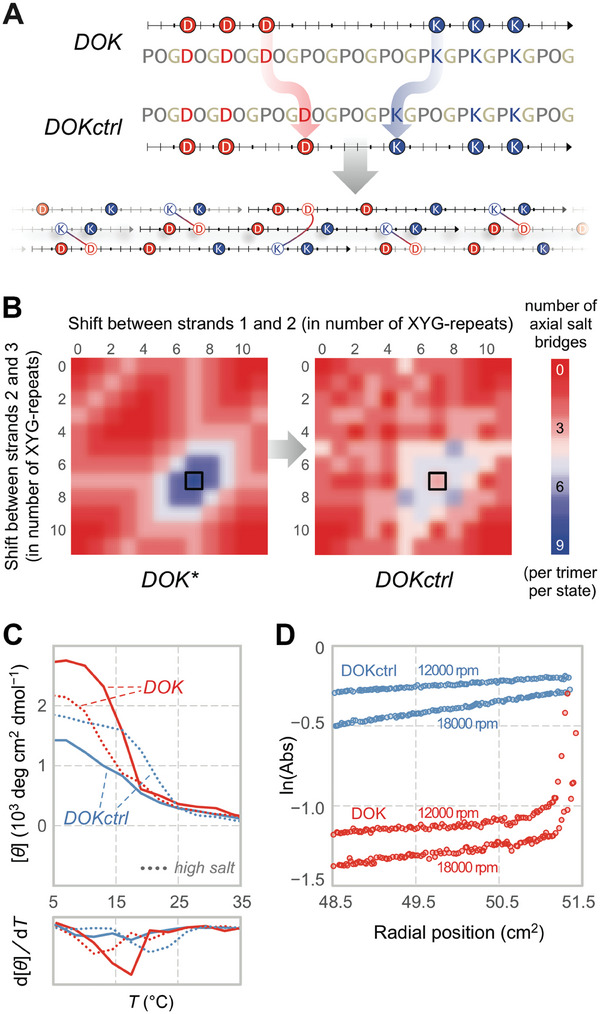
Disruption of the salt‐bridge network eliminates symmetric self‐assembly. A) A slight modification to the DOK sequence results in a peptide (DOKctrl) that is unable to establish ⅔ of the salt bridges accessible to its parent upon symmetric assembly. This change is reflected in the strand‐association landscape (SAL) for DOKctrl B). For a given CMP, SAL presents the number of axial salt bridges supported by every possible strand association leading to sticky‐ended self‐assembly into an extended triple helix. The symmetric association state (SAS; black borders) appears at tripeptide shifts of (7,7), which is equivalent to (−4,−4), and provides 9 and 3 axial salt bridges per trimer for DOK and DOKctrl, respectively. DOKctrl loses the well‐defined singular maximum of DOK at the SAS. *This landscape describes strand association for all 11/3sb permutants, as well as DOK. C) CD thermal denaturation curves for DOK and DOKctrl (0.6 mg/mL) at low (20 mm, solid line) and high ionic strength (200 mm, dotted line). D) Sedimentation equilibrium gradients for DOK and DOKctrl (0.3 mg/mL). The divergent impact of ionic strength on triple‐helicity and differences in assembly size both indicate dissimilar molecular organization when strand association is disrupted.
