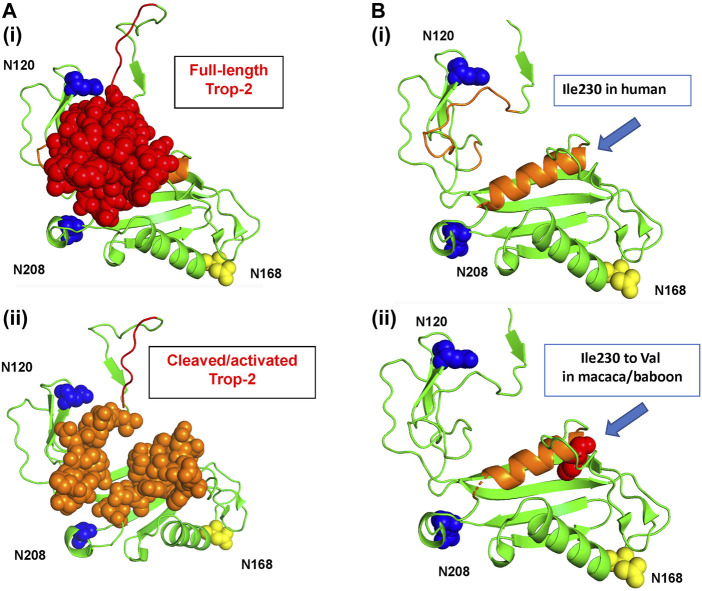
FIGURE 1.
The human Trop-2 protein structure. The 3D structure of the extra-cellular domain of Trop-2 (Pavšič, 2021) is in ribbon diagrams and sphere/protein surface models. Trop-2 N-glycosylation sites are indicated. N120 and N-208 are in blue; the N120A and N-208A mutants were shown to abolish binding of Claudin-7 to Trop-2 (Mori et al., 2019; Kamble et al., 2021). The N168 glycosylation site is in yellow. (A) Top view. (i) The N-terminal subunit of Trop-2 is in red as sphere model; (ii) the 3D structure of the extra-cellular domain of Trop-2 devoid of the N-terminal subunit is shown. The groove between the glycans at N120 and N208 that becomes more accessible upon ADAM10-cleavage and rearrangement of the N-terminal ADAM10-cleaved subunit (Guerra et al., 2023a) is in orange, sphere model. (B) Ribbon diagrams of the activation site of Trop-2 (Mori et al., 2019; Kamble et al., 2021), and binding site of Hu2G10 (Guerra et al., 2023a) are provided for clarity. Blue arrow: polymorphic residue across NHP species. (i) Human Ile230 residue. (ii) Space-fill model of cynomolgus and baboon Val230 is provided. The residue is buried at the bottom of the orange α-helix, below the solvent-exposed region.