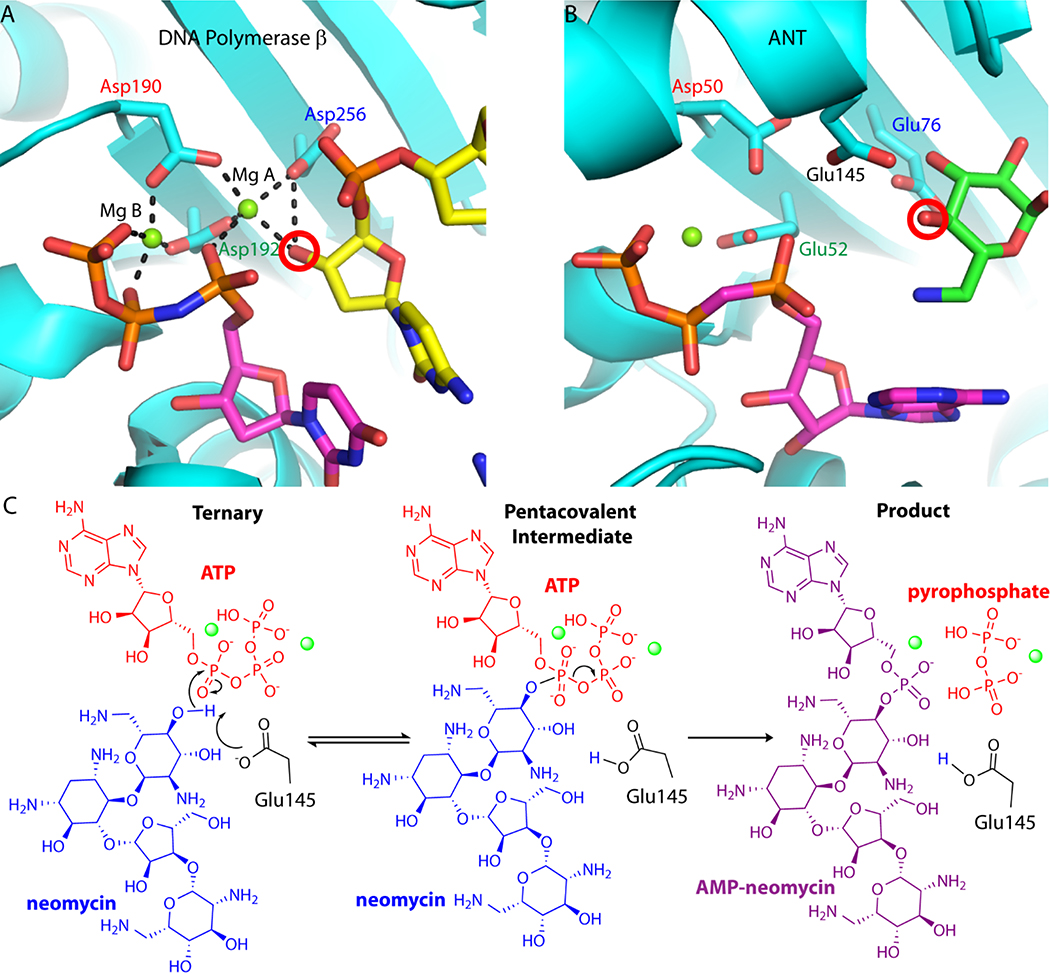
Figure 1.
Conservation of nucleotidyl transfer active sites. A. Close-up view of the Pol b ternary complex active site (PDB code 2FMS). Protein is colored in cyan, the NTP is shown with magenta carbon atoms, and the DNA primer terminus is shown with yellow carbon atoms. In Pol b, the nucleotide binding metal (Mg B) is ligated with the b and g phosphates of the nucleotide triphosphate, as well as Asp190 and Asp192. The catalytic metal (Mg A) is coordinated by all three aspartate residues, including the catalytic base (Asp256) and the activated nucleophile hydroxyl from the carbohydrate moiety. Hydrogen bonds are represented as black dashed lines. B. Close-up view of the D80Y ANT4’ ternary complex active site (PDB code 1KNY). Protein is colored in cyan, nucleotide substrate is shown with magenta carbon atoms, and kanamycin is shown with carbon atoms colored green. Conserved active site carboxylate groups of D80Y ANT4’ and Pol b are shown and labelled in the same color. The nucleophilic oxygen on neomycin is circled in red. In ANT4’ Asp50 and Glu52 are homologous to Asp190 and Asp192 in Pol b. C. The proposed nucleotidyl transfer reaction of ANT4’. The Mg2+ ions involved in the reaction coordinate are represented as green spheres. In these studies, the far left diagram was captured crystallo-graphically (ATP was replaced with non-hydrolyzable AMCPCPP) as well as the far right diagram.