Abstract
PD-1+TCF-1+ stem-like CD8 T cells act as critical resource cells for maintaining T cell immunity in chronic viral infections and cancer. In addition, they provide the proliferative burst of effector CD8 T cells after PD-1 directed immunotherapy. An important unanswered question is whether the numbers of these stem-like progenitor cells are diminished in this process of enhanced effector differentiation. We have addressed this question using the mouse model of chronic LCMV infection. We found that treatment of chronically infected mice with either αPD-1 or αPD-L1 not only increased effector cell differentiation from the virus-specific stem-like CD8 T cells, but also increases their proliferation so their numbers were maintained. This increased self-renewal of LCMV-specific stem-like CD8 T cells was mTOR-dependent. We did microscopy studies to better understand the division of these progenitor cells after PD-1 blockade and found that an individual dividing cell could give rise to a differentiated TCF-1− daughter cell alongside a self-renewing TCF-1+ sister cell. This asymmetric division also helps in preserving the number of stem-like cells. We then examined whether the gene expression program and functionality of the PD-1+TCF-1+ stem-like CD8 T cells were modified after the PD-1 blockade. We found that the stem-like CD8 T cells retained their transcriptional program and also their in vivo functionality in terms of responding to viral infection and to repeat PD-1 blockade. Taken together, our results demonstrate that PD-1 blockade does not deplete the stem-like population despite increasing effector differentiation. These findings have implications for PD-1 directed immunotherapy in humans.
One Sentence Summary:
PD-1 regulates not only effector differentiation from PD-1+TCF-1+ stem-like CD8 T cells but also their self-renewal.
INTRODUCTION
It is now well established that antigen-specific PD-1+TCF-1+ stem-like CD8 T cells play a major role in sustaining CD8 T cell responses during chronic viral infections and cancer (1–6). These quiescent stem-like CD8 T cells, also referred to as precursors of exhausted CD8 T cells, do not express effector molecules and reside in lymphoid tissues where they undergo a slow self-renewal and can also proliferate and differentiate into effector CD8 T cells that eventually get exhausted (7–14). Most importantly, PD-1 directed immunotherapy acts specifically on this subset of cells—inducing a proliferative burst that accelerates their differentiation into effectors (12–14). This substantial increase in the numbers of effector CD8 T cells emerging from the stem-like cells after removing the PD-1 brake, combined with PD-1 blockade at the target site, results in efficient killing of virally-infected or tumor cells (14–18). PD-1 is now the leading immune check point inhibitor and PD-1 directed immunotherapy is approved for the treatment of several different cancers (19–24).
A critical question that is not well understood is what happens to the numbers of PD-1+TCF-1+ stem-like CD8 T cells after PD-1 therapy? It is not known if there is a loss of the pool of these progenitor cells as they undergo increased effector differentiation. Moreover, we also do not know if the gene expression program or functionality of the stem-like CD8 T cells is changed after PD-1 directed immunotherapy. We have addressed these questions in this study and investigated the impact of PD-1 therapy on virus-specific TCF-1+ stem-like CD8 T cells during chronic LCMV infection of mice (25–27).
RESULTS
Virus-specific stem-like CD8 T cell numbers are maintained following PD-1 blockade
Mice chronically infected with LCMV (>day 45) were treated with αPD-1 or αPD-L1 antibody for two weeks and then examined for CD8 T cell responses in the spleen (Fig. S1 and S2A). As in earlier studies, PD-1 blockade resulted in an increase in the percentage and numbers of PD-1+ and LCMV-specific CD8 T cells, based on staining with H-2DbGP33 and H-2DbGP276 tetramers (Fig. S2B, C, and D) (28). Also consistent with previous studies, the largest increase both in terms of the percentage and total numbers was seen in the PD-1+Tim-3+CX3CR1+CD101− transitory effector CD8 T cells that are derived from the stem-like CD8 T cells (Fig. S2E, F, and G) (14, 15). Consequently, there was a decrease in the percentage of the PD-1+TCF-1+ stem-like CD8 T cells (Fig. 1B–G). However, there was no decrease in the total numbers of these progenitor cells (Fig. 1H, I, and J). In fact, there was an increase in the numbers of the TCF-1+ stem-like cells after PD-1 blockade; this was seen for total PD-1+ progenitor cells and for LCMV-specific GP33+ and GP276+ progenitor CD8 T cells (Fig. 1H, I, and J). This was surprising, and may not have been observed in previous studies where quantitation of the stem-like CD8 T cell subset after PD-1 blockade had not been performed extensively. The data shown in Figure 1 are based on a large number of mice (n=54 to 75), thus providing good statistical significance. An increase in TCF-1+ stem-like cells was generally observed across multiple independent experiments (Fig. S3). In a few experiments, the increase in the number of progenitor CD8 T cells did not reach statistical significance, but in these instances, there was still no trend towards a decrease in their numbers. We also examined the stem-like CD8 T cell response after PD-1 blockade in the LCMV Clone 13 chronic infection model without transient CD4 T cell depletion and saw the same results; the numbers of stem-like cells increased, or were at least maintained, following PD-1 blockade in both GP276+ and GP33+ populations (Fig. S4). Ultimately, we found that stem-like CD8 T cells were not only maintained at day 14 following PD-1 blockade, but their population also remained stable for another 8 weeks in the absence of further treatment (Fig. 2).
Fig. 1. PD-1 blockade decreases the frequency of virus-specific stem-like CD8 T cells but their numbers increase.
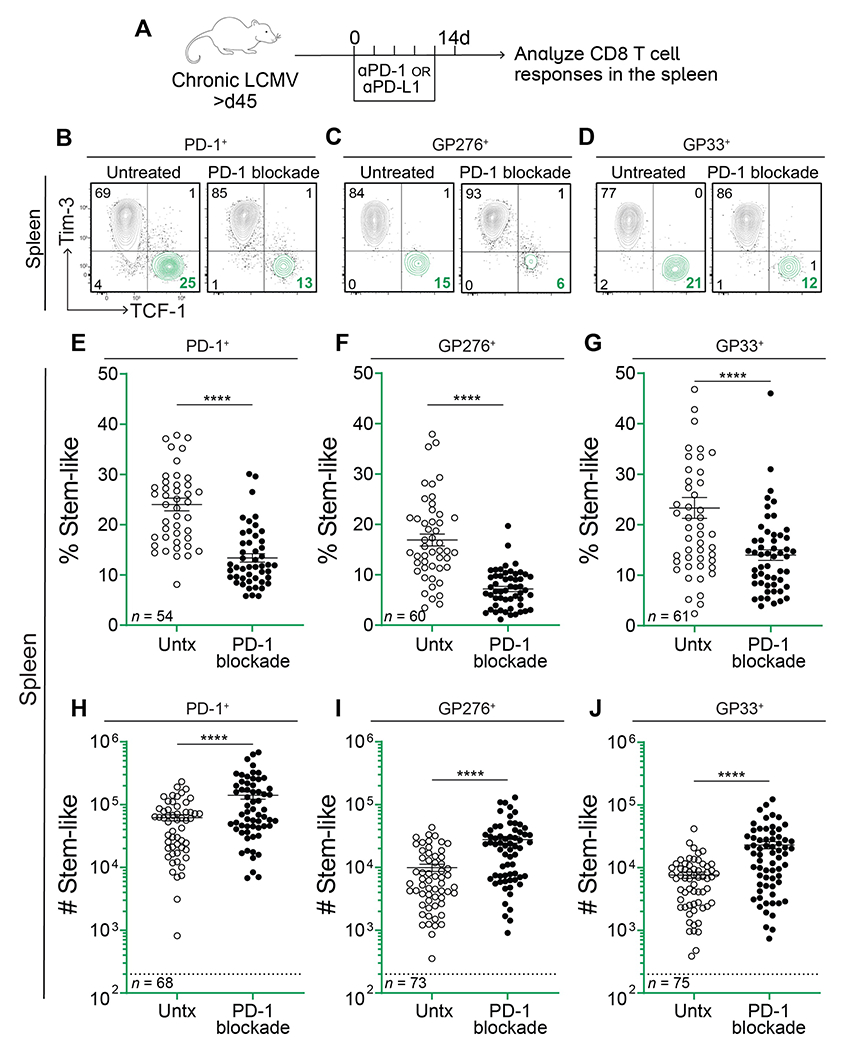
(A) LCMV chronically infected mice were treated with αPD-1 or αPD-L1 every 3 days for 2 weeks. CD8 T cell responses were analyzed on day 14. (B-D) Representative flow plots showing expression of TCF-1 and Tim-3 on total PD-1+, GP276+, and GP33+ CD8 T cells with and without αPD-1/L1 treatment. (E-G) Frequency of TCF-1+Tim-3− stem-like cells among the LCMV-specific CD8 T cell populations indicated above each plot. (H-J) Absolute number of stem-like cells among the LCMV-specific populations indicated above each plot. Dotted lines indicate the limit of detection. The number of mice in each plot is shown in the bottom left corner and values are from 12 independent experiments. Individual data points represent individual mice; bars represent mean ± SEM. Statistical significance in (E-J) was determined using unpaired two-tailed t tests; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****; P ≤ 0.0001; ns, non-significant.
Fig. 2. Virus-specific stem-like CD8 T cells are maintained up to 8 weeks following initial PD-1 blockade.
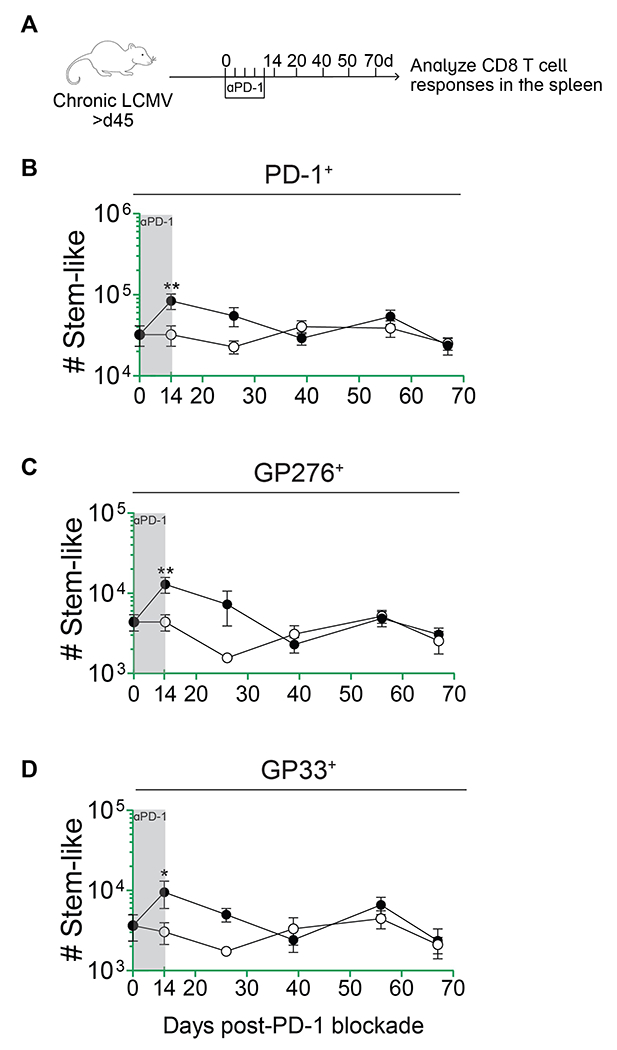
(A) LCMV chronically infected mice were treated with αPD-1 every 3 days for 2 weeks. Following treatment cessation, CD8 T cell responses were analyzed at weeks 2, 4, 6, 8, and 10. (B-D) Absolute number of stem-like cells among the LCMV-specific populations indicated above each plot. Data points represent means from 3 pooled experiments ± SEM; n=3-5 mice per group per timepoint; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****; P ≤ 0.0001; ns, non-significant.
Virus-specific stem-like CD8 T cells increase their self-renewal following PD-1 blockade
As stem-like CD8 T cells did not decrease in numbers while PD-1 blockade was actively recruiting them to differentiate, we hypothesized that PD-1 blockade may contribute to their maintenance by increasing their proliferation. To address this, we examined the proliferation of stem-like CD8 T cells at days 8 and 14 after PD-1 blockade. After 8 days of treatment, the total number of stem-like CD8 T cells was significantly higher as compared to the untreated group (P=0.0039 among PD-1+ stem-like cells; Fig. S5D), and we found that this expanded compartment also expressed higher levels of Ki67 (Fig. 3). There was an increase in the percentage and numbers of Ki67+PD-1+TCF-1+ CD8 T cells at day 8 after PD-1 blockade (Fig. 3A, D, and G). The same pattern was seen with LCMV-specific PD-1+TCF-1+ stem-like CD8 T cells (Fig. 3B–C, E–F, and H–I). At day 14 after PD-1 blockade, the percentage of Ki67+ stem-like CD8 T cells had returned to baseline levels (Fig. 3D–F), but the total numbers of proliferating cells were still higher (Fig. 3G–I). These results show that PD-1 blockade not only promotes the differentiation of stem-like CD8 T cells into effector cells but it also increases the proliferation and self-renewal of the stem-like cells themselves. This ensures that the numbers of these critical resource CD8 T cells are maintained following PD-1 blockade.
Fig. 3. Increased proliferation and self-renewal of virus-specific stem-like CD8 T cells after PD-1 blockade.
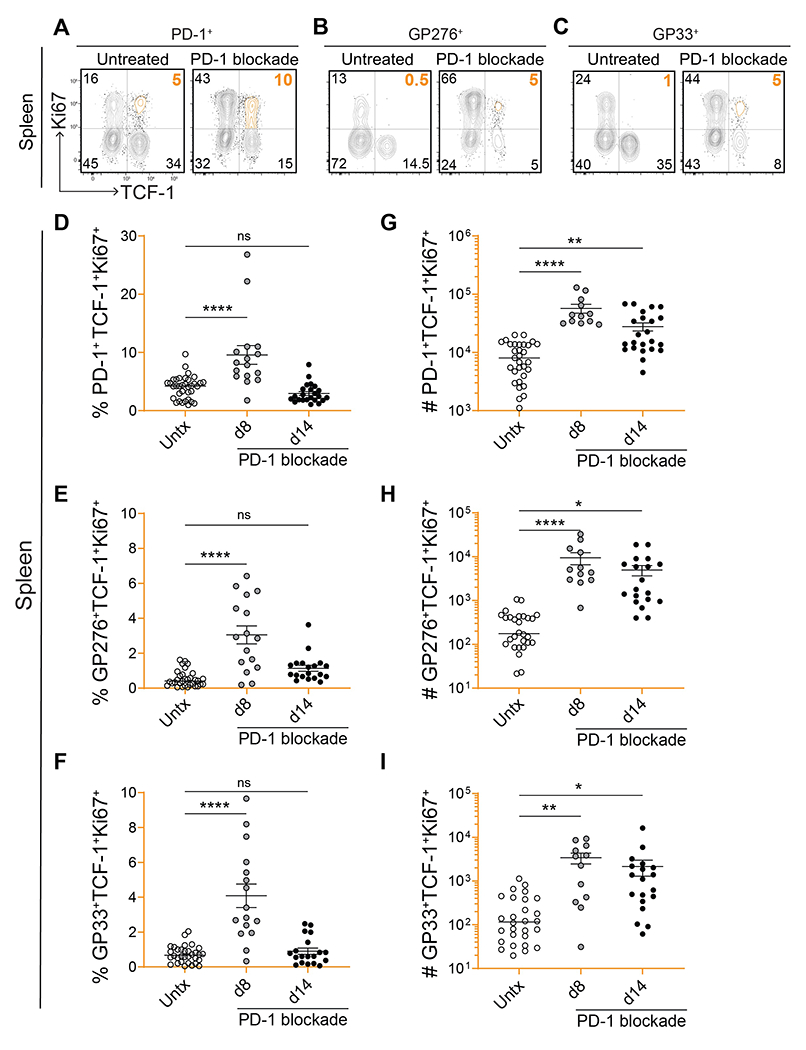
(A-C) Representative flow plots showing expression of TCF-1 and Ki67 on total PD-1+, GP276+, and GP33+ CD8 T cells on day 8 post-PD-1 blockade. (D-I) Frequency and absolute number of stem-like PD-1+, GP276+, or GP33+ CD8 T cells that were proliferating (Ki67+) on days 8 and 14 post-blockade. Individual data points represent individual mice; bars represent mean ± SEM; 4-7 independent experiments per group, n=3-5 mice per experiment. Statistical significance (D-I) was calculated using one-way ANOVA with Tukey’s test for multiple comparisons; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****; P ≤ 0.0001; ns, non-significant.
mTOR signaling is needed for increased proliferation and self-renewal of virus-specific stem-like CD8 T cells after PD-1 blockade
Earlier studies have shown that mTOR signaling is required for effector CD8 T cell proliferation and differentiation from the stem-like CD8 T cells following PD-1 blockade during chronic LCMV infection (29, 30). So, we next asked if mTOR signaling was also needed for increased self-renewal of virus-specific stem-like CD8 T cells after blockade of the PD-1 inhibitory pathway. We administered rapamycin to chronically infected mice daily for a period of 8 days or 14 days, in combination with αPD-L1 antibody treatment (Fig. 4A). Control mice received sham treatment during the same time period. Following αPD-L1 treatment, the number of stem-like CD8 T cells increased, as previously shown, but the addition of rapamycin abrogated this effect, limiting their numbers to pre-treatment levels (Fig. 4B and S6). Examination on day 8 showed that the absolute number of proliferating Ki67+TCF-1+ stem-like CD8 T cells was also reduced following the addition of rapamycin (Figs. 4C and S7). Thus, mTOR signaling is needed for stem-like cells’ increased self-renewal and the PD-1 inhibitory pathway also regulates this process.
Fig. 4. mTOR signaling regulates proliferation of virus-specific stem-like CD8 T cells after PD-1 therapy.
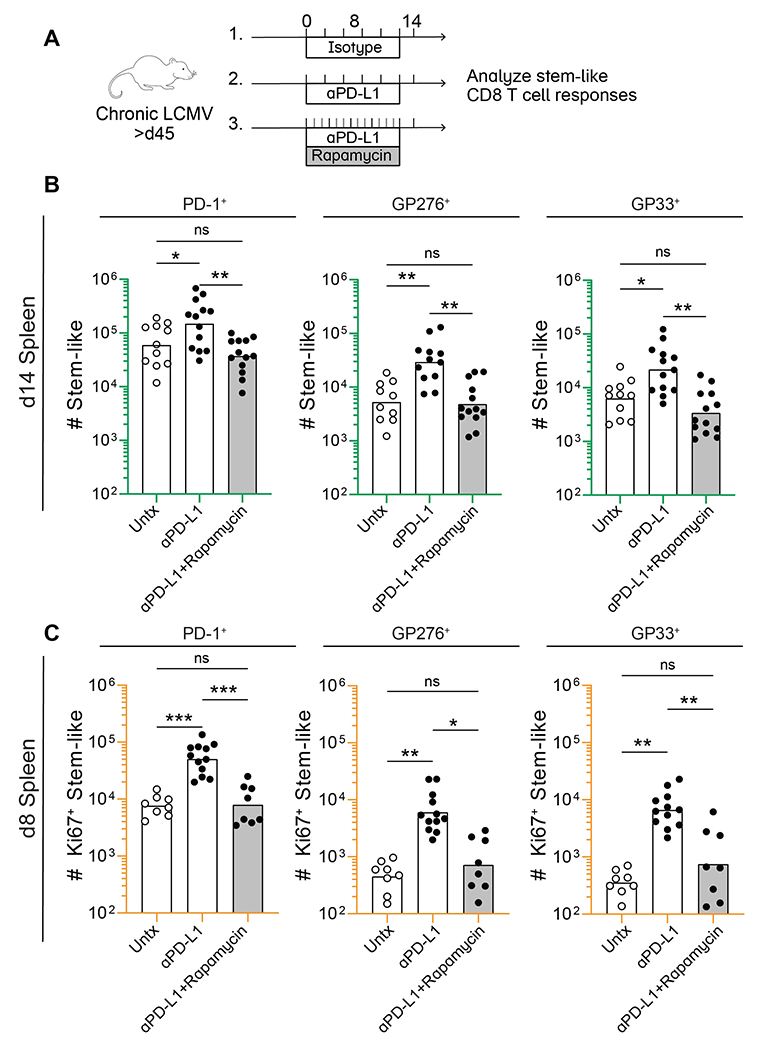
(A) Rapamycin was administered to mice daily in combination with αPD-L1 for 8 or 14 days. Control mice received sham treatment during the same time period. (B) Absolute number of stem-like CD8 T cells in the spleen among total PD-1+, GP276+, and GP33+ CD8 T cells following 2 week treatment with isotype, αPD-L1, or αPD-L1 in combination with rapamycin (C) Absolute number of proliferating (Ki67+) stem-like CD8 T cells among the total LCMV-specific populations indicated above each plot on day 8 post-treatment. (B) and (C) each show values from 3 independent experiments, n=3-5 mice per group, per experiment. Individual data points represent individual mice, and bars indicate group means. Statistical significance (B and C) was calculated using one-way ANOVA with Tukey’s test for multiple comparisons; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****; P ≤ 0.0001; ns, non-significant.
Stem-like CD8 T cells can undergo asymmetric division to maintain their numbers
We investigated whether stem-like CD8 T cells might be programmed to undergo cellular divisions that promote maintenance of their numbers following PD-1 blockade. Specifically, we sought to determine whether stem-like CD8 T cells might utilize asymmetric cell division to allocate a differentiated daughter cell alongside another self-renewing, stem-like sibling cell. Thus, we performed immunofluorescence microscopy on dividing progenitor cells from untreated and αPD-L1-treated mice at day 8 post-treatment (Fig. 5A). Confocal microscopy of singlet CD8 T cells revealed readily detectable expression of PD-1 and TCF-1 (Fig. 5B). Conjoined sibling pairs undergoing cytokinesis were identified by the presence of a tubulin bridge connecting two adjacent cell bodies (Fig. 5C). We analyzed PD-1+CD8+ conjoined sibling cell pairs that contained at least one TCF-1+ cell and observed both concordant division (two TCF-1 positive daughters) and discordant division (one TCF-1 positive daughter and one TCF-1 negative daughter) indicating self-renewal versus differentiation, respectively (Fig. 5D). Both types of division were detectable in untreated and αPD-L1-treated stem-like cells. Of note, concordant TCF-1− daughters were also present and may represent progeny from either a TCF-1+ or TCF-1− parent. Altogether, these results suggest that stem-like progenitor CD8 T cells can achieve their own self-renewal alongside continued production of differentiated effector cells through asymmetric cell division, either during steady-state or under conditions of enhanced effector output following PD-1 blockade.
Fig. 5. Asymmetric division can sustain the stem-like CD8 T cell population.
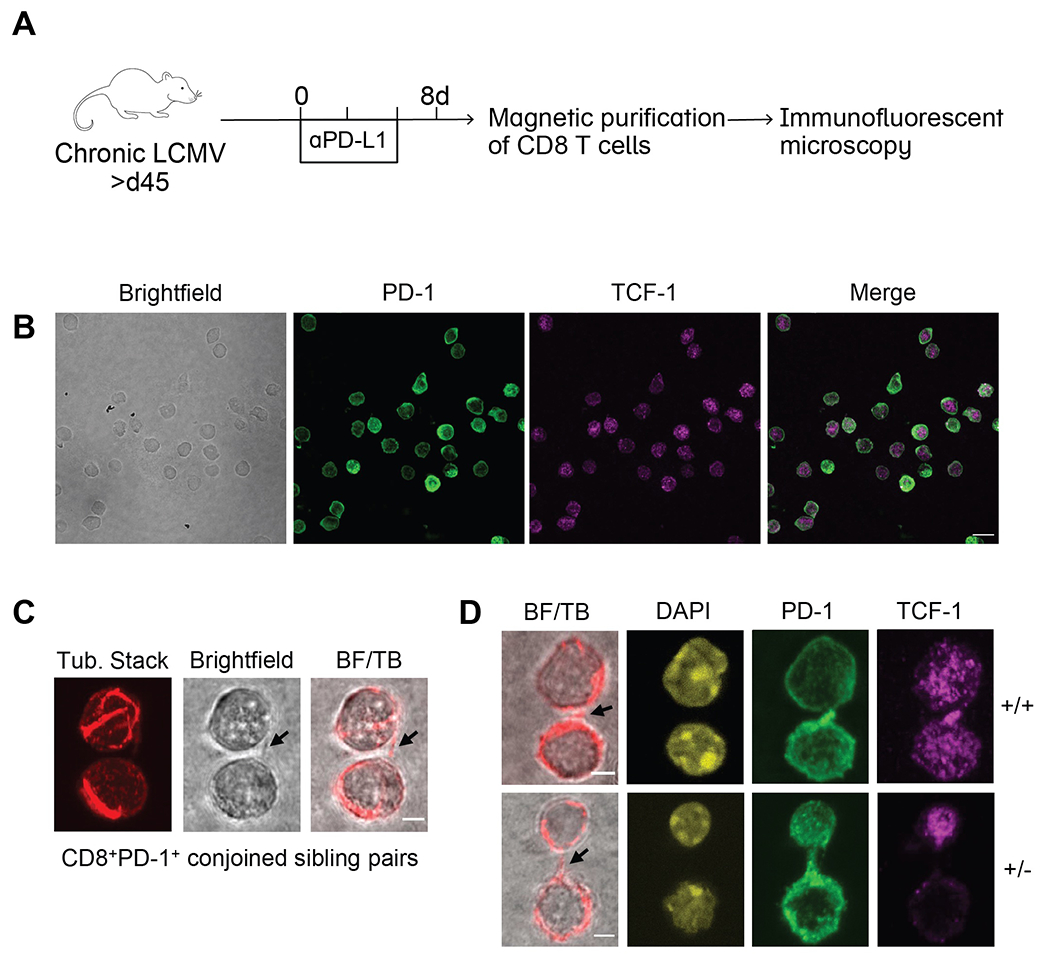
(A) LCMV chronically infected mice were treated with or without αPD-L1 every 3 days. CD8 T cells were isolated from spleens by magnetic purification on day 8 and immunofluorescence confocal microscopy was performed. (B) Representative photos of singlet CD8 T cells following αPD-L1 treatment were captured on 60X lens with 2X scanning zoom (0.1μm/pixel). Merged fluorescence of PD-1 and TCF-1 are shown. Scale bar is 10μm. (C) Conjoined CD8+PD-1+ sibling pairs were captured on 60X lens with 4X scanning zoom (0.05 μm/pixel). Representative conjoined sibling cell pair illustrating bridge between sibling cells marked by arrows in (l-to-r) brightfield and merge of brightfield with single z-slice tubulin (BF/TB). (D) CD8+PD-1+ sibling pairs from αPD-L1-treated mice illustrating TCF-1 concordant (+/+; top) and TCF-1 discordant (+/−; bottom) divisions. (−/−) sibling pairs were also observed but are not shown. Representative images displaying (l-to-r), brightfield merge with single z-slice tubulin (bridges marked by arrows), DAPI, PD-1, and TCF-1 staining (n=14 doublets imaged per group). Scale bars are 2.5μm. (B) and (C) each show representative images from a total of 3 independent experiments.
The transcriptional program of stem-like CD8 T cells is conserved following PD-1 blockade
We next investigated whether stem-like CD8 T cells undergo functional or transcriptional changes following an extensive proliferative history, as after PD-1 therapy. Previous studies have demonstrated transcriptional and epigenetic stability of exhausted CD8 T cells following immunotherapy (31, 32), but we sought to study the effect of PD-1 blockade on the stem-like progenitor population specifically. To do this, we sorted stem-like CD8 T cells and the more differentiated Tim-3+ CD8 T cells from LCMV chronically infected mice that had been treated with αPD-1 antibody for 14 days or from chronically infected mice that had received no treatment (Fig. S8A). RNA-sequencing (RNA-seq) analysis was done on these sorted CD8 T cell subsets from the two groups of mice. We found that the treated stem-like CD8 T cells were transcriptionally almost identical to untreated stem-like cells. Using principal component analysis (PCA), we compared the subsets and treatment groups: sorted stem-like cells clustered together tightly along both PC1 and PC2—regardless of prior treatment (Fig. S8B). A heatmap of selected genes also showed minimal to no changes in the stem-like CD8 T cells after PD-1 blockade (Fig. S8C). We further confirmed these findings by doing single cell RNA-seq (scRNA-seq) on LCMV GP33 tetramer sorted CD8 T cells from untreated or PD-1 treated chronically infected mice (Fig. S8D, E, and F). This dataset was originally generated by Hashimoto et al., but we performed additional analyses on the stem-like compartment (17). At day 14 post-treatment, the frequency of proliferating Ki67+ cells by flow cytometry had essentially returned to baseline (Fig. 3). Therefore, closer to the peak of proliferation, we may have uncovered more heterogeneity in gene expression between the treatment groups, for example, among cell cycle or metabolic genes. Thus, our data are consistent with the transcriptional programming of stem-like cells being conserved following PD-1 therapy, and after several rounds of cell proliferation.
Virus-specific stem-like CD8 T cells retain their functionality after PD-1 blockade
We next asked whether the functionality of stem-like CD8 T cells remained intact after the first cycle of PD-1 therapy. To address this question we examined whether the virus-specific PD-1+TCF-1+stem-like CD8 T cells retained the ability to proliferate and differentiate following a viral challenge and whether they could respond efficiently to a second round of PD-1 blockade.
In the first set of experiments, we transferred stem-like cells from untreated or cycle 1 αPD-1-treated chronically infected mice into naïve recipients and challenged these mice with LCMV Clone 13 (Fig. 6A). Robust proliferation of donor stem-like CD8 T cells was observed in the blood between days 0 and 14 post-infection, with equivalent expansion of untreated stem-like cells and PD-1 treated stem-like cells (Fig. 6B, right). As expected, there was minimal expansion of the more differentiated Tim-3+PD-1+ CD8 T cells (Fig. 6B, left). A similar pattern was seen in the spleen, liver, and lungs at day 14 after infection, with stem-like CD8 T cells from both groups of mice expanding equally well (Fig. 6C–I). Phenotypically, donor stem-like cells underwent classical differentiation to become Tim-3+TCF-1−, while a small proportion remained Tim-3−TCF-1+ (Fig. 6J). Thus, no defect was observed in the proliferative capacity of cycle 1-treated versus untreated stem-like cells in any tissue examined.
Fig. 6. Stem-like CD8 T cells that have received cycle 1 PD-1 therapy can robustly proliferate in response to viral challenge.
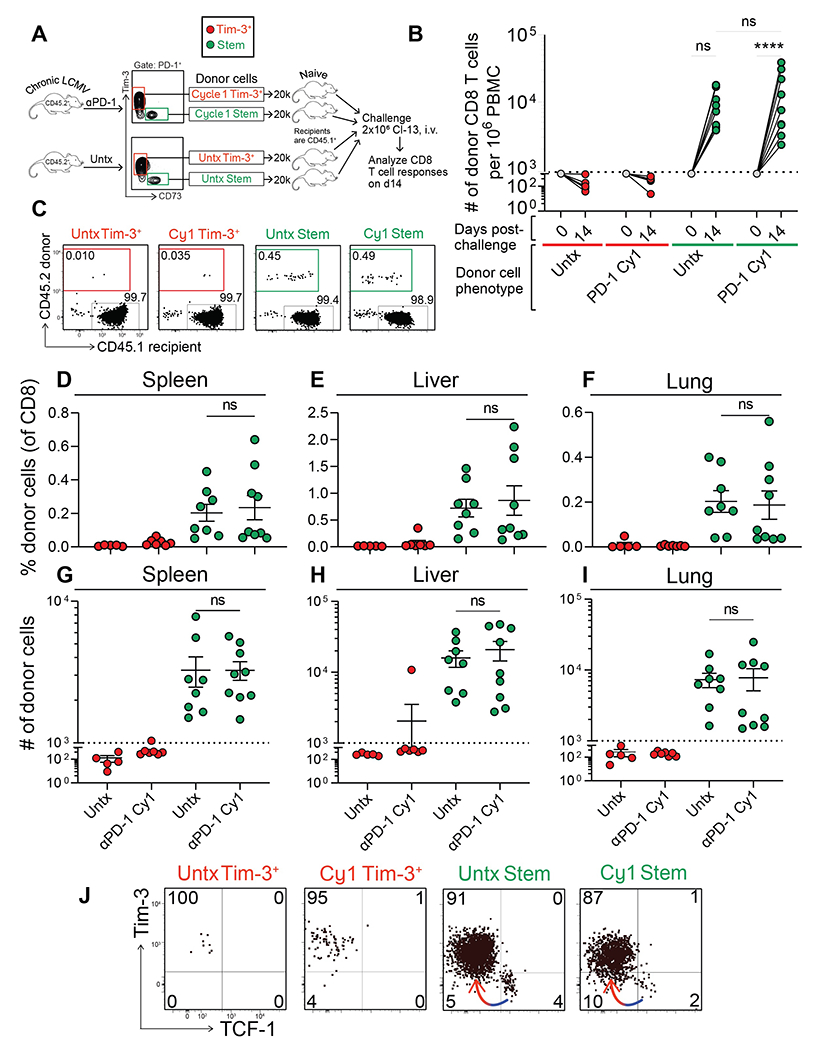
(A) Stem-like (PD-1+Tim-3−CD73+) and Tim-3+ (PD-1+Tim-3+CD73−) CD8 T cells were each sorted from isotype or cycle 1 αPD-1-treated CD45.2+ donor mice. These subsets were transferred into naïve CD45.1+ recipients that were challenged with LCMV Clone 13 (2 × 106 pfu, i.v.) the next day. Donor CD8 T cell responses were analyzed on day 14 post-challenge. (B) Expansion of donor cells among PBMC at day 14 post-challenge. Dotted line indicates the limit of detection based on 5% estimated take of donor cells within the recipient. (C) Frequency of donor (CD45.2+) and recipient (CD45.1+) CD8 T cells in the spleen. Treatment of the donor cells prior to transfer is shown above each plot. Frequency (D-F) or absolute number (G-I) of donor CD8 T cells within the spleen, liver, or lung. (J) Phenotype of donor CD8 T cells based on Tim-3 and TCF-1 expression. Colored arrows indicate the direction of differentiation. Two independent transfer experiments were performed, each with n=2-4 mice per group. In all plots, individual data points represent individual mice; bars represent mean ± SEM. Two-way Statistical significance was determined by ANOVA with Sidak’s test for multiple comparisons (B) or one-way ANOVA with Tukey’s test for multiple comparisons (D-I); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****; P ≤ 0.0001; ns, non-significant.
As αPD-1 treatment did not dampen the magnitude of proliferation or the differentiation of stem-like CD8 T cells in response to Clone 13 challenge, we next investigated whether αPD-1-treated stem-like cells responded to secondary PD-1 blockade. We isolated cycle 1 αPD-1-treated stem-like cells, transferred them into infection-matched, untreated recipients, and treated them with a second round of αPD-1 (or isotype control) (Fig. 7A). We found that cycle 1-treated stem-like cells were able to mount a robust proliferative response following an additional cycle of PD-1 blockade, as compared to the cycle 1-treated stem-like cells that did not receive a second round of therapy—by both frequency and total number of cells in the PBMC, spleen, liver, and lungs (Fig. 7B–G). Responses were equivalent to those from cycle 1 blockade alone (Fig. S9). Furthermore, cycle 1-treated stem-like cells retained their capacity to seed a pool of Tim-3+TCF-1− effector cells without becoming depleted themselves (Fig. 7H and I, as shown by the arrow).
Fig. 7. Stem-like CD8 T cells that have received cycle 1 PD-1 therapy can respond to an additional cycle.
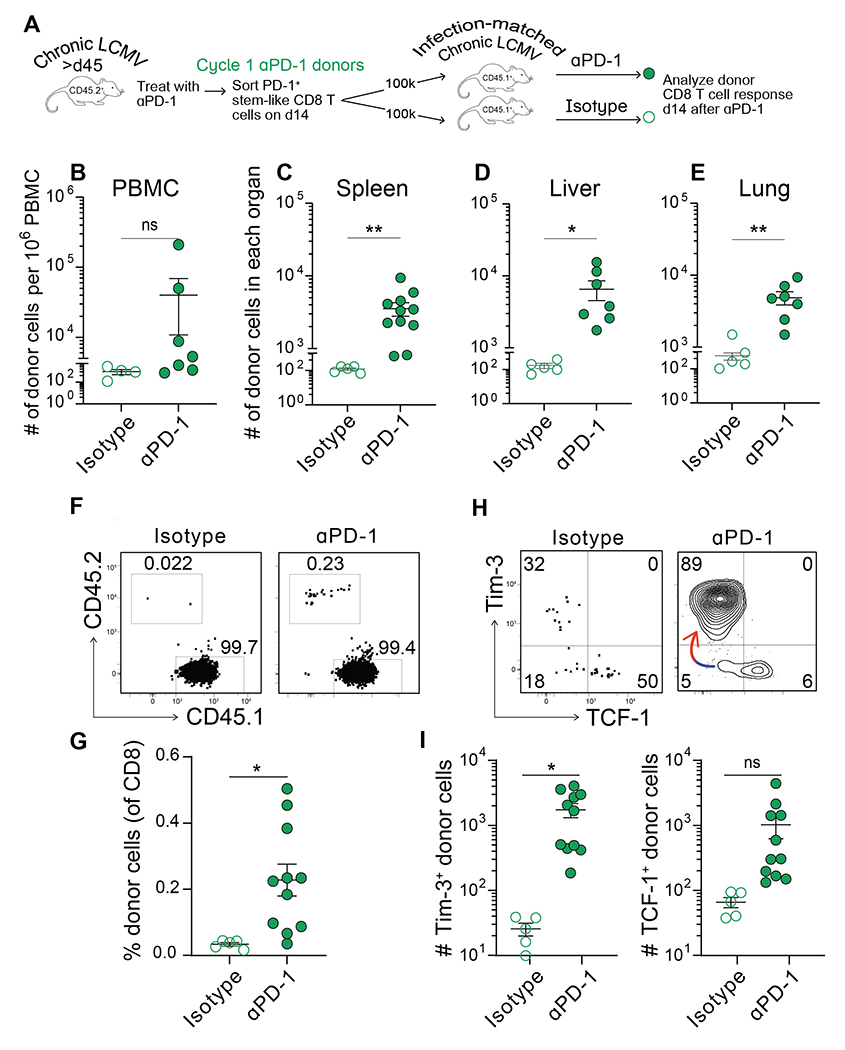
(A) Stem-like CD8 T cells were sorted from cycle 1 αPD-1-treated CD45.2+ donor mice. These cells were transferred into infection-matched CD45.1+ recipients that were then treated with an additional round of αPD-1. Donor CD8 T cell responses were analyzed on day 14. (B-E) Absolute number of donor (CD45.2+) CD8 T cells within the PBMC, spleen, liver, or lung at day 14 post-secondary PD-1 blockade. (F) Frequency of donor (CD45.2+) and recipient (CD45.1+) CD8 T cells as a proportion of transferred cells following the recipient’s treatment (αPD-1 or isotype). (G) Frequency of donor CD8 T cells following the recipient’s treatment (H) Phenotype of the donor CD8 T cell population after additional treatment, based on Tim-3 and TCF-1 expression. Colored arrow indicates the direction of differentiation. (I) Absolute number of Tim-3+TCF-1− and Tim-3−TCF-1+ donor CD8 T cells after additional treatment. Two independent transfer experiments were performed, each with n=2-4 mice per group. In all plots, individual data points represent individual mice; bars represent mean ± SEM. Statistical significance in (B-E, G, and I) was determined using unpaired two-tailed t tests; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****; P ≤ 0.0001; ns, non-significant.
DISCUSSION
These studies using the mouse model of chronic LCMV infection have addressed a critical and unanswered question about what happens to the numbers and function of PD-1+TCF-1+ stem-like CD8 T cells after PD-1 therapy. It is now well established that PD-1 blockade increases the proliferation and differentiation of stem-like CD8 T cells into effector cells (12–14) and a potential concern was whether this accelerated differentiation would lead to a loss of the stem-like CD8 T cells. Our study now shows that PD-1 blockade also increases the self-renewal of virus-specific stem-like CD8 T cells thereby compensating for any potential loss and preventing the depletion of this important precursor population (Fig. 8). We also show that the transcriptional signature and functionality of the stem-like CD8 T cells are retained after PD-1 blockade.
Fig. 8. CD8 T cell differentiation during chronic infection and after PD-1 blockade.
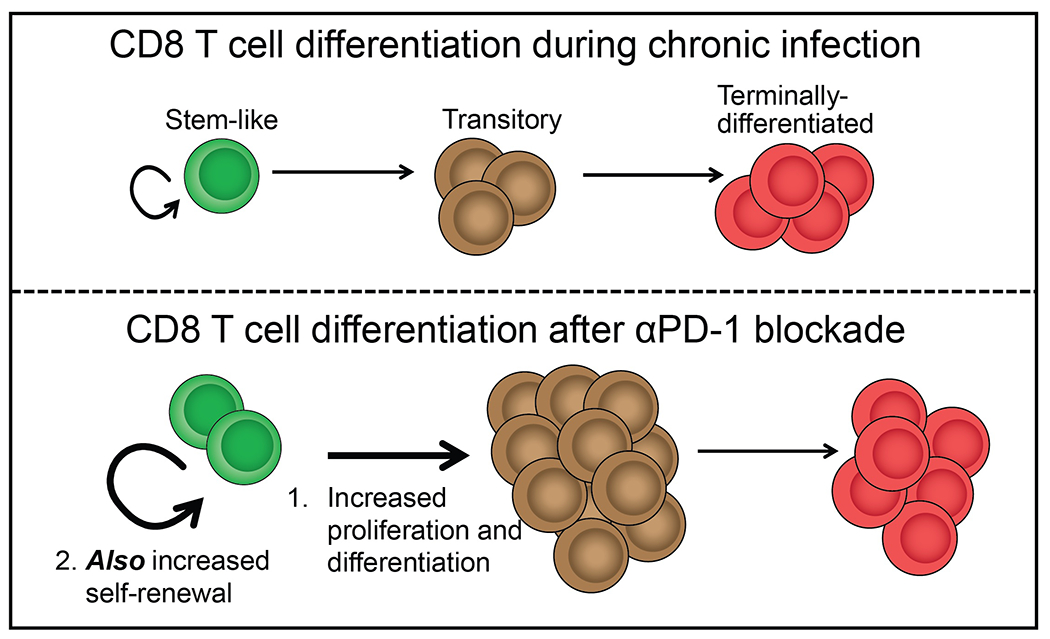
At steady state during chronic infection, stem-like CD8 T cells undergo slow self-renewal and steadily turn over into transitory effector CD8 T cells. These cells sustain the antiviral response, but gradually become exhausted. Following PD-1 blockade, stem-like CD8 T cells undergo a proliferative burst to increase the pool of effector cells, but they also increase their self-renewal, ensuring that they are not depleted.
We found that stem-like cells can undergo asymmetric cell division. By microscopy, PD-1+TCF-1+ stem-like CD8 T cells were able to produce a differentiated TCF-1 negative daughter cell alongside a self-renewing TCF-1 positive sibling cell. This would allow the number of stem-like cells to be maintained, whilst the number of effector cells is simultaneously increasing following PD-1 blockade. This asymmetric cell division of the PD-1+ stem-like CD8 T cells was seen in both untreated chronically infected mice and after PD-1 blockade. This is a simple and elegant mechanism for maintaining the numbers of stem-like CD8 T cells during chronic infection.
The PD-1 inhibitory receptor negatively regulates TCR signaling and also signaling by costimulatory molecules (33–35). The major effect of PD-1 blockade is increased generation of effector CD8 T cells from the stem-like CD8 T cells. It is interesting that our studies now show that the self-renewal of the stem-like CD8 T cells is also regulated by the PD-1 inhibitory pathway. In other words, PD-1 is playing a role in regulating two types of proliferation; proliferation resulting in differentiation into effectors and also proliferation associated with self-renewal of the stem-like CD8 T cells. We found that mTOR signaling is required for the proliferation of stem-like cells following PD-1 blockade. Previously, Ando et al. used rapamycin to demonstrate the requirement of mTOR signaling in the generation of effector-like CD8 T cells following PD-1 blockade (29). Thus, both types of proliferation are mTOR dependent. Several different signals including TCR and costimulation can activate the mTOR pathway (36–38). Future studies are needed to study the signals that regulate effector differentiation versus self-renewal of the stem-like CD8 T cells after PD-1 directed immunotherapy. An interesting study in this issue of Science Immunology shows that CD28 co-stimulation plays a role in both effector differentiation and self-renewal of stem-like CD8 T cells during chronic LCMV infection and in tumor models (39).
Finally, we show that stem-like CD8 T cells remain transcriptionally and functionally intact following PD-1 blockade. After two weeks of PD-1 therapy, the stem-like CD8 T cells maintain their stemness at the transcriptional level and can respond to viral infection and a second cycle of PD-1 therapy. However, it needs to be determined if this will hold up when PD-1 therapy is continued for extended periods of time.
In conclusion, our studies have addressed a key question about the potential loss of the PD-1+TCF-1+ stem-like CD8 T cells following PD-1 blockade. Our studies show that this is prevented by increased self-renewal of these cells and also by the ability of these cells to undergo asymmetric division thus maintaining their numbers. It will be important to extend these studies to other model systems and to see if this increased self-renewal is also seen in human chronic infections and cancer after PD-1 therapy.
MATERIALS AND METHODS
Study Design
The objective of this study was to investigate the effect of PD-1 blockade on maintenance and functionality of the stem-like CD8 T cell population. For this, we evaluated (i) the number, (ii) the gene expression profile, and (iii) the in vivo proliferative capacity of stem-like cells from chronically infected mice after PD-1 therapy. These studies were complemented by further analysis of the mechanisms underlying increased proliferation and self-renewal in the setting of PD-1 therapy. We performed experiments using rapamycin, an inhibitor of the mTOR pathway, to evaluate the role of mTOR signaling during stem-like cells’ proliferation. Additionally, we examined cellular divisions by immunofluorescent microscopy to evaluate the role of asymmetric division in maintaining the stem-like population. All mouse experiments were performed with 3- to 12 biological replicates per experimental group. All experiments were repeated at least twice unless otherwise stated.
Mice and viruses.
LCMV infections were performed as previously described (12). All animal experiments were performed in accordance with the Emory University Institutional Animal Care and Use Committee. C57BL/6J and CD45.1 congenic female mice were purchased from Jackson Laboratory. For chronic infections, 6-8 week old mice were injected intraperitoneally (i.p.) with 300 μg of the CD4 T cell-depleting antibody GK1.5 (Bio X Cell) one day before and one day after intravenous (i.v.) injection with 2 × 106 pfu LCMV Clone 13. One experiment was also performed without this transient CD4 depletion (Fig. S3, as indicated in the figure legend).
PD-1 blockade.
LCMV chronically infected mice (>45 days post-infection) received either PD-1 blockade or no treatment. Mice that received PD-1 blockade were injected i.p. with either 200μg of mouse anti-mouse αPD-1 (332.8H3, clone 2203, mouse IgG1 with D265A mutation in the Fc portion) or 200μg of rat anti-mouse αPD-L1 (clone 10F.9G2; in house). Control mice received 200μg of isotype control in PBS or no injection. Mice were injected every 3 days and sacrificed 8 or 14 days after the first dose.
Administration of rapamycin.
Rapamycin (Wyeth) was administered to mice i.p. daily for 8 days or 2 weeks. The daily dose of rapamycin was 600 μg kg−1 (blood levels; 40–100 ng ml−1), diluted in 1% Tween 80 and 2% ethanol in Phosal 50 PG (40). Control mice received daily sham treatment (injection of the buffer without rapamycin).
Cell isolation.
Spleens were isolated from mice and smashed through 70μm nylon cell strainers to remove clumps. Cells were spun down at 2,000 RPM for 10 minutes, and the supernatant was removed. After resuspending the pellet, cells were incubated at room temperature with 1 ml of Ammonium-Chloride-Potassium (Ack) lysis buffer for 2 minutes in order to lyse the RBCs. After incubation, an additional 35 ml of RPMI + 5% FBS was added to dilute the Ack lysis buffer, and the cells were spun down at 2,000 RPM for 10 minutes. The resulting cell pellet was resuspended in PBS + 2% FBS + 0.5mM EDTA and cells were transferred to a 96-well round-bottom plate for staining. For lungs, tissue was incubated at 37°C with Collagenase-I prior to straining. For both the liver and lungs, lymphocytes were isolated via Percoll gradient.
Flow cytometry.
Surface staining was performed by incubating cells with fluorochrome-conjugated antibodies against CD8 (clone 53-6.7; PerCP-eFluor 710 from eBioscience; BUV395 or BUV737 from BD Bioscience), CD4 (clone GK1.5; APC-Cy7 from Biolegend; BUV661 from BD Bioscience), CD19 (clone 1D3; APC-Cy7 from Biolegend; BUV661 from BD Bioscience), PD-1 (clone 29F.1A12; PE and BV785 from Biolegend), CD44 (clone IM7; APC-R700 from Biolegend; BUV737 from BD Bioscience), Tim-3 (clone RMT3-23; BV421 from Biolegend), CD101 (clone Moushi101; PE-Cy7 from eBioscience), CX3CR1 (clone SA011F11; BV605 from Biolegend), CD45.1 (clone A20; BUV395 from BD Bioscience), and CD45.2 (clone 104; FITC from Biolegend). Cells were incubated on ice for 30 minutes in PBS + 2% FBS + 0.5mM EDTA. Ki67 (clone B56; FITC, BV711, or BUV395 from BD Bioscience) and TCF-1 (clone C63D9 from Cell Signaling Technology, followed by goat anti-rabbit AF488 secondary from Invitrogen) were stained intracellularly with the eBioscience Foxp3/Transcription Factor Fixation/Permeabilization Kit (ThermoFisher). For detecting LCMV-specific CD8 T cell responses, peptides bound to major histocompatibility complex (pMHC) tetramers were prepared against LCMV GP33 and LCMV GP276 proteins, as described previously (41, 42). Cell viability was determined with the Live/Dead fixable aqua or near IR dead cell stain kit (Invitrogen). Samples were acquired on an LSR II or Symphony flow cytometer (BD Bioscience) and data were analyzed with FlowJo software (TreeStar).
Bulk RNA-sequencing and analysis.
PD-1+Tim-3+CD73− (terminally-differentiated) and PD-1+Tim-3−CD73+ (stem-like) CD8 T cells were sorted using Fluorescence-Activated Cell Sorting (FACS) on day 14 following αPD-1 blockade or isotype treatment. The following fluorochrome-conjugated antibodies were used for the sort: CD4 (clone GK1.4; APC-Cy7 from Biolegend), CD19 (clone 1D3; APC-Cy7 from Biolegend), CD8 (clone 53-6.7; APC from Biolegend), PD-1 (clone RMP1-30; PE from Biolegend), Tim-3 (clone 215008; Alexa Fluor 488 from R and D systems), and CD73 (clone TY/11.8; PE-Cy7 from Biolegend). Naive CD44loCD62Lhi CD8 T cells from uninfected mice were also isolated by FACS using CD44 (clone IM7; FITC from BD Bioscience) and CD62L (clone MEL-14; PE from Biolegend). Samples were sorted on a FACSAria II (BD Bioscience). After sorting, RNA was isolated from each sample using the All-Prep DNA/RNA Micro Kit (Qiagen) and then submitted to the Emory Yerkes Nonhuman Primate Genomics Core. Reads from RNA-seq were aligned to the mm10 genome [accessed through Ensembl (43)] with STAR version 2.7. The DESeq2 package [v1.32.0 (44)] was used to detect differentially expressed genes and plots were made with ggplot2 or GraphPad Prism.
Single cell RNA-sequencing and analysis.
scRNA-seq data was obtained from Hashimoto et al., deposited at GEO under accession code GSE206739 (17). In their study, mice infected with LCMV Clone 13 for >45 days were treated with αPD-L1 or left untreated. On day 14 post-treatment, mice were sacrificed and GP33+ CD8+ T cells were magnetically enriched and then sorted on a FACS Aria II (BD Bioscience). The initial study included additional treatment groups, but we only re-analyzed the sequencing from the naïve, untreated, and αPD-L1 groups. The Yerkes Nonhuman Primate Genomics Core generated a single cell gene expression library, and this data was aligned using CellRanger version 4 (10X Genomics). Outlier cells with high numbers of reads originating from mitochondrial genes and presumed doublets were excluded from the dataset. Genes encoded on mitochondrial chromosomes were also excluded from analysis. Data was normalized and scaled using the Seurat package [v4.0.4 (45–48)] and plots were made with ggplot2 or GraphPad Prism. Shared nearest neighbor clustering was performed in Seurat with 200 neighbors, 6 principal components (1-7, excluding principal component 4, which was primarily based on ribosomal genes), and a resolution of 0.8. UMAP dimensionality reduction was performed with identical parameters and a minimum distance of 2. A phylogenetic tree was also created with identical parameters, using Seurat’s BuildClusterTree. For analysis of the stem-like cluster individually, prior clustering was used to generate a new data object, on which additional UMAP dimensionality reduction was performed (with 500 neighbors, principal components 1-7 [excluding 4], a resolution of 0.9, and a minimum distance of 1).
Adoptive transfer experiments.
For the transfer followed by LCMV challenge: Mice infected with LCMV Clone 13 (as above) for >45 days were injected i.p. with αPD-1 or IgG1 isotype control in PBS. Injections were given every 3 days and mice were sacrificed on day 14 and pooled into αPD-1-treated or isotype-treated groups. CD8 T cells in the samples were purified with the EasySep Mouse CD8 T Cell Isolation Kit (StemCell Technologies), then two CD8 T cell subsets were sorted from each treatment group using FACS: PD-1+Tim-3+CD73− (terminally-differentiated) and PD-1+Tim-3−CD73+ (stem-like). Cells were reconstituted in RPMI and 2-2.5 × 104 sorted cells from each group were transferred i.v. into uninfected, age-matched CD45.1+ recipients. On the next day, the recipients were challenged with 2 × 106 pfu LCMV Clone 13, i.v. Recipients were bled one week post-transfer, and then sacrificed 2 weeks post-transfer, at which time cells were isolated from the spleen, liver, lung, and PBMC, and stained as above.
For the transfer followed by secondary PD-1 blockade: Mice infected with LCMV Clone 13 (as above) for >45 days were injected with αPD-1 every 3 days. On day 14, mice were sacrificed and isolated splenocytes were pooled, purified with the EasySep Mouse CD8 T Cell Isolation Kit (StemCell Technologies), and sorted using FACS into a pure population of PD-1+Tim-3−CD73+ stem-like CD8 T cells. Sorted cells were reconstituted in RPMI and 1 × 105 cells were transferred i.v. into infection-matched CD45.1+ recipients. Recipients were then treated with a second cycle of αPD-1 therapy and sacrificed 2 weeks later. At this time, cells were isolated from the spleen, liver, lung, and PBMC, and stained as above. In some experiments, isotype-treated donor stem-like CD8 T cells were also isolated, sorted, and transferred, treated with a secondary blockade.
Confocal immunofluorescent microscopy.
Chronically infected mice (>45 days post-infection) received no treatment or αPD-L1 every 3 days for 3 doses. On day 8, mice were sacrificed and isolated splenocytes were purified with the EasySep Mouse CD8 T Cell Isolation Kit (StemCell Technologies). Purified CD8 T cells were shipped overnight on ice in RPMI with 10% FBS to Columbia University Irving Medical Center for immunofluorescence staining as previously described (49). Briefly, 50,000 cells were transferred to each coverslip (ThermoFisher) coated with poly-l-lysine (Sigma-Aldrich) and allowed to adhere at 37°C before 4% paraformaldehyde fixation (Electron Microscopy Sciences), quenching with 50mM NH4Cl (Sigma-Aldrich) in PBS and 0.1% triton X-100 in PBS (Sigma-Aldrich), then blocking overnight with blocking buffer comprised of 0.25% fish skin gelatin (Sigma-Aldrich) and 0.01% saponin (Sigma-Aldrich) in PBS. The following day, cells were stained with rat anti-beta-tubulin (clone YOL1/34; ThermoFisher supplier Novus Biologicals), mouse anti-PD-1 (332.8H3, clone 2203; in house), and rabbit anti-TCF-1 (C63D9; Cell Signaling Technology) for 1 hour at room temperature, followed by blocking buffer washes. Subsequently, secondary antibody staining with goat anti-rat 488 (ThermoFisher), goat anti-mouse 568 (ThermoFisher), and goat anti-rabbit 647 (ThermoFisher) was performed for 1 hour at room temperature, followed by blocking buffer washes and water rinses. ProLong Diamond Antifade Mountant with DAPI (ThermoFisher) was used to mount stained cells to frosted microscope slides (ThermoFisher) overnight. Nail polish was used to seal coverslips to microscope slides the following day.
Images were collected at the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University. Z-stacked images were acquired on a Nikon Ti Eclipse laser-scanning inverted confocal microscope 60x/1.49 Apo TIRF oil lens (Nikon) with 405nm, 488nm, 561nm, and 638nm lasers. Transmitted light and some tubulin images were from a single-z plane. Other tubulin images, DAPI, PD-1, and TCF-1 were displayed by creating a maximum projection image from a z-stack summary. Image analysis was performed by Fiji V2.3.0/1.53f (National Institutes of Health). Sibling cells undergoing cytokinesis exhibited a cytoplasmic cleft between cell bodies under transmitted light, with a defined tubulin bridge within the cleft, plus dual nuclei DAPI staining. Assessment of sibling status was performed blinded to the staining of TCF-1. Comparative quantitation of TCF-1 between sibling cells was calculated by taking the (fluorescence of daughter 1 – fluorescence of daughter 2)/ (fluorescence of daughter 1 + fluorescence of daughter 2), with values over 0.2 considered asymmetric. Sibling pairs were verified by tubulin bridge. Sibling pairs with PD-1 intensity in the top 75th percentile were considered PD-1+.
Statistical analysis.
All experiments were analyzed using Prism 9 (GraphPad Software). Statistical differences were assessed using unpaired two-tailed t tests, one-way ANOVA with Tukey’s test for multiple comparisons, or two-way ANOVA with Sidak’s or Dunnett’s tests for multiple comparisons.
Supplementary Material
Acknowledgments:
We thank Robert Karaffa and Kametha Fife for assistance with cell sorting in the Emory University School of Medicine Flow Cytometry Core, and Kathryn Pellegrini for assistance with bulk RNA-seq out of the Yerkes Nonhuman Primate Genomics Core. We thank Gordon Freeman for generously donating his mouse Fc-mutated αPD-1 antibody. We also thank Wendy Lin, Dylan Mariuzza, Robert Washburn, Theresa Swayne, and Haojie Ji for assistance with imaging at Columbia University.
Funding:
This work was supported by NIH grants R01AI030048 (to RA), NIH grants R01CA279268, R56AI076458, T32 P30CA013696, R01AI106711, and V Foundation (to SLR and PHW), NIH grant R00AI153736 (to WHH), NIH grant R01AI39675 (to KA), NIH grant P01 AI056299 (to GJF), HHMI Hanna Gray Fellowship GT16001 (to CBM), Burroughs Wellcome Fund Postdoctoral Enrichment Program 1022361 (to CBM), and Achievement Rewards for College Scientists Standard Scholarship (to ALG). The Yerkes Nonhuman Primate Genomics Core is supported by NIH grant P51OD011132.
Footnotes
Competing interests: RA and GJF hold patents on the PD-1 pathway. GJF has served on advisory boards for Roche, Bristol-Myers-Squibb, iTeos, NextPoint, IgM, Jubilant, GV20, IOME, Santa Ana Bio, Simcere of America, and Geode. The other authors declare that they have no competing interests.
Data and materials availability:
All RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE223399. Mouse anti-mouse αPD-1 (332.8H3, clone 2203, mouse IgG1 with D265A mutation in the Fc portion) can be provided by GJF pending a completed material transfer agreement. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Brummelman J, Mazza EMC, Alvisi G, Colombo FS, Grilli A, Mikulak J, Mavilio D, Alloisio M, Ferrari F, Lopci E, Novellis P, Veronesi G, Lugli E, High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. Journal of Experimental Medicine. 215, 2520–2535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eberhardt CS, Kissick HT, Patel MR, Cardenas MA, Prokhnevska N, Obeng RC, Nasti TH, Griffith CC, Im SJ, Wang X, Shin DM, Carrington M, Chen ZG, Sidney J, Sette A, Saba NF, Wieland A, Ahmed R, Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature. 597, 279–284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L, Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 537, 412–416 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, Zotos D, Fenix KA, Atnerkar A, Preston S, Chipman JG, Beilman GJ, Allison CC, Sun L, Wang P, Xu J, Toe JG, Lu HK, Tao Y, Palendira U, Dent AL, Landay AL, Pellegrini M, Comerford I, McColl SR, Schacker TW, Long HM, Estes JD, Busslinger M, Belz GT, Lewin SR, Kallies A, Yu D, CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol. 17, 1187–1196 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, Cardenas M, Wilkinson S, Lake R, Sowalsky AG, Valanparambil RM, Hudson WH, McGuire D, Melnick K, Khan AI, Kim K, Chang YM, Kim A, Filson CP, Alemozaffar M, Osunkoya AO, Mullane P, Ellis C, Akondy R, Im SJ, Kamphorst AO, Reyes A, Liu Y, Kissick H, An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 576, 465–470 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, Luther SA, Speiser DE, Held W, Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity. 50, 195–211.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, Johnson J, Staupe RP, Bengsch B, Xu C, Yu S, Kurachi M, Herati RS, Vella LA, Baxter AE, Wu JE, Khan O, Beltra J-C, Giles JR, Stelekati E, McLane LM, Lau CW, Yang X, Berger SL, Vahedi G, Ji H, Wherry EJ, TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity (2019), doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T, Ji Y, Moseman EA, Xu HC, Manglani M, Kirby M, Anderson SM, Handon R, Kenyon E, Elkahloun A, Wu W, Lang PA, Gattinoni L, McGavern DB, Schwartzberg PL, The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Science Immunology. 1, eaai8593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jadhav RR, Im SJ, Hu B, Hashimoto M, Li P, Lin J-X, Leonard WJ, Greenleaf WJ, Ahmed R, Goronzy JJ, Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. PNAS, 201903520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Im SJ, Konieczny BT, Hudson WH, Masopust D, Ahmed R, PD-1+ stemlike CD8 T cells are resident in lymphoid tissues during persistent LCMV infection. PNAS (2020), doi: 10.1073/pnas.1917298117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dähling S, Mansilla AM, Knöpper K, Grafen A, Utzschneider DT, Ugur M, Whitney PG, Bachem A, Arampatzi P, Imdahl F, Kaisho T, Zehn D, Klauschen F, Garbi N, Kallies A, Saliba A-E, Gasteiger G, Bedoui S, Kastenmüller W, Type 1 conventional dendritic cells maintain and guide the differentiation of precursors of exhausted T cells in distinct cellular niches. Immunity (2022), doi: 10.1016/j.immuni.2022.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, Sharpe AH, Freeman GJ, Germain RN, Nakaya HI, Xue H-H, Ahmed R, Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utzschneider DT, Charmoy M, Chennupati V, Pousse L, Ferreira DP, Calderon-Copete S, Danilo M, Alfei F, Hofmann M, Wieland D, Pradervand S, Thimme R, Zehn D, Held W, T Cell Factor 1-Expressing Memory-like CD8+ T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin J-X, Konieczny BT, Im SJ, Freeman GJ, Leonard WJ, Kissick HT, Ahmed R, Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity. 51, 1043–1058.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zander R, Schauder D, Xin G, Nguyen C, Wu X, Zajac A, Cui W, CD4+ T Cell Help Is Required for the Formation of a Cytolytic CD8+ T Cell Subset that Protects against Chronic Infection and Cancer. Immunity. 51, 1028–1042.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang PH, Washburn R, Maniar R, Mu M, Ringham O, Kratchmarov R, Henick BS, Reiner SL, Cutting Edge: Promoting T Cell Factor 1+ T Cell Self-Renewal to Improve Programmed Cell Death Protein 1 Blockade. The Journal of Immunology (2022), doi: 10.4049/jimmunol.2200317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto M, Araki K, Cardenas MA, Li P, Jadhav RR, Kissick HT, Hudson WH, McGuire DJ, Obeng RC, Wieland A, Lee J, McManus DT, Ross JL, Im SJ, Lee J, Lin J-X, Hu B, West EE, Scharer CD, Freeman GJ, Sharpe AH, Ramalingam SS, Pellerin A, Teichgräber V, Greenleaf WJ, Klein C, Goronzy JJ, Umaña P, Leonard WJ, Smith KA, Ahmed R, PD-1 combination therapy with IL-2 modifies CD8+ T cell exhaustion program. Nature. 610, 173–181 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Codarri Deak L, Nicolini V, Hashimoto M, Karagianni M, Schwalie PC, Lauener L, Varypataki EM, Richard M, Bommer E, Sam J, Joller S, Perro M, Cremasco F, Kunz L, Yanguez E, Hüsser T, Schlenker R, Mariani M, Tosevski V, Herter S, Bacac M, Waldhauer I, Colombetti S, Gueripel X, Wullschleger S, Tichet M, Hanahan D, Kissick HT, Leclair S, Freimoser-Grundschober A, Seeber S, Teichgräber V, Ahmed R, Klein C, Umaña P, PD-1-cis IL-2R agonism yields better effectors from stem-like CD8+ T cells. Nature. 610, 161–172 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribas A, Wolchok JD, Cancer immunotherapy using checkpoint blockade. Science. 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM, Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. New England Journal of Medicine. 366, 2455–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJM, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ, Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng S, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ, MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 515, 558–562 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn M-J, Felip E, Lee J-S, Hellmann MD, Hamid O, Goldman JW, Soria J-C, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L, Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N Engl J Med. 372, 2018–2028 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Hamid O, Robert C, Daud A, Hodi FS, Hwu W-J, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, Dronca R, Gangadhar TC, Patnaik A, Zarour H, Joshua AM, Gergich K, Elassaiss-Schaap J, Algazi A, Mateus C, Boasberg P, Tumeh PC, Chmielowski B, Ebbinghaus SW, Li XN, Kang SP, Ribas A, Safety and Tumor Responses with Lambrolizumab (Anti–PD-1) in Melanoma. N Engl J Med. 369, 134–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallimore A, Glithero A, Godkin A, Tissot AC, Plückthun A, Elliott T, Hengartner H, Zinkernagel R, Induction and Exhaustion of Lymphocytic Choriomeningitis Virus–specific Cytotoxic T Lymphocytes Visualized Using Soluble Tetrameric Major Histocompatibility Complex Class I–Peptide Complexes. The Journal of Experimental Medicine. 187, 1383–1393 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R, Viral Persistence Alters CD8 T-Cell Immunodominance and Tissue Distribution and Results in Distinct Stages of Functional Impairment. Journal of Virology. 77, 4911–4927 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJD, Suresh M, Altman JD, Ahmed R, Viral Immune Evasion Due to Persistence of Activated T Cells Without Effector Function. The Journal of Experimental Medicine. 188, 2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R, Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 439, 682–687 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Ando S, Perkins C, Sajiki Y, Chastain C, Valanparambil RM, Wieland A, Hudson WH, Hashimoto M, Ramalingam SS, Freeman GJ, Ahmed R, Araki K, mTOR regulates T cell exhaustion and PD-1 targeted immunotherapy response during chronic viral infection. J Clin Invest (2022), doi: 10.1172/JCI160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabriel SS, Tsui C, Chisanga D, Weber F, Llano-León M, Gubser PM, Bartholin L, Souza-Fonseca-Guimaraes F, Huntington ND, Shi W, Utzschneider DT, Kallies A, Transforming growth factor-β-regulated mTOR activity preserves cellular metabolism to maintain long-term T cell responses in chronic infection. Immunity. 54, 1698–1714.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao H-W, Godec J, LaFleur MW, Brown FD, Tonnerre P, Chung RT, Tully DC, Allen TM, Frahm N, Lauer GM, Wherry EJ, Yosef N, Haining WN, The epigenetic landscape of T cell exhaustion. Science. 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, Barnitz RA, Bartman C, Bengsch B, Huang AC, Schenkel JM, Vahedi G, Haining WN, Berger SL, Wherry EJ, Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharpe AH, Pauken KE, The diverse functions of the PD1 inhibitory pathway. Nature Reviews Immunology. 18, 153–167 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD, T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science. 355, 1428–1433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, Sica GL, Sharpe AH, Freeman GJ, Blazar BR, Turka LA, Owonikoko TK, Pillai RN, Ramalingam SS, Araki K, Ahmed R, Rescue of exhausted CD8 T cells by PD-1–targeted therapies is CD28-dependent. Science. 355, 1423–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, Collins SL, Lutz MA, Allen AN, Kole TP, Zarek PE, Powell JD, A Role for Mammalian Target of Rapamycin in Regulating T Cell Activation versus Anergy. The Journal of Immunology. 178, 2163–2170 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Long L, Zhou P, Chapman NM, Chi H, mTOR signaling at the crossroads of environmental signals and T-cell fate decisions. Immunological Reviews. 295, 15–38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson AW, Turnquist HR, Raimondi G, Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol. 9, 324–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humblin Etienne, Korpas Isabel, Lu Jiahua, Filipescu Dan, van der Heide Verena, Goldstein Simon, Vaidya Abishek, Soares-Schanoski Alessandra, Casati Beatrice, Selvan Myvizhi Esai, Gümüş Zeynep H., Wieland Andreas, Corrado Mauro, Cohen-Gould Leona, Bernstein Emily, Homann Dirk, Chipuk Jerry, Kamphorst Alice O., Sustained CD28 costimulation is required for self-renewal and differentiation of TCF-1+ PD-1+ CD8 T cells. Science Immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson RP, Hartman DA, Ochalski SJ, Zimmerman JL, Glaser KB, Sirolimus (rapamycin, Rapamune®) and combination therapy with cyclosporin A in the rat developing adjuvant arthritis model: Correlation with blood levels and the effects of different oral formulations. Inflamm. res 47, 339–344 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM, Phenotypic Analysis of Antigen-Specific T Lymphocytes. Science. 274, 94–96 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJD, Zajac AJ, Miller JD, Slansky J, Ahmed R, Counting Antigen-Specific CD8 T Cells: A Reevaluation of Bystander Activation during Viral Infection. Immunity. 8, 177–187 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, Berry A, Bhai J, Bignell A, Billis K, Boddu S, Brooks L, Charkhchi M, Cummins C, Da Rin Fioretto L, Davidson C, Dodiya K, Donaldson S, El Houdaigui B, El Naboulsi T, Fatima R, Giron CG, Genez T, Martinez JG, Guijarro-Clarke C, Gymer A, Hardy M, Hollis Z, Hourlier T, Hunt T, Juettemann T, Kaikala V, Kay M, Lavidas I, Le T, Lemos D, Marugán JC, Mohanan S, Mushtaq A, Naven M, Ogeh DN, Parker A, Parton A, Perry M, Piližota I, Prosovetskaia I, Sakthivel MP, Salam AIA, Schmitt BM, Schuilenburg H, Sheppard D, Pérez-Silva JG, Stark W, Steed E, Sutinen K, Sukumaran R, Sumathipala D, Suner M-M, Szpak M, Thormann A, Tricomi FF, Urbina-Gómez D, Veidenberg A, Walsh TA, Walts B, Willhoft N, Winterbottom A, Wass E, Chakiachvili M, Flint B, Frankish A, Giorgetti S, Haggerty L, Hunt SE, IIsley GR, Loveland JE, Martin FJ, Moore B, Mudge JM, Muffato M, Perry E, Ruffier M, Tate J, Thybert D, Trevanion SJ, Dyer S, Harrison PW, Howe KL, Yates AD, Zerbino DR, Flicek P, Ensembl 2022. Nucleic Acids Res. 50, D988–D995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R, Integrated analysis of multimodal single-cell data. Cell. 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satija R, Farrell JA, Gennert D, Schier AF, Regev A, Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin W-HW, Adams WC, Nish SA, Chen Y-H, Yen B, Rothman NJ, Kratchmarov R, Okada T, Klein U, Reiner SL, Asymmetric PI3K Signaling Driving Developmental and Regenerative Cell Fate Bifurcation. Cell Reports. 13, 2203–2218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE223399. Mouse anti-mouse αPD-1 (332.8H3, clone 2203, mouse IgG1 with D265A mutation in the Fc portion) can be provided by GJF pending a completed material transfer agreement. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.


