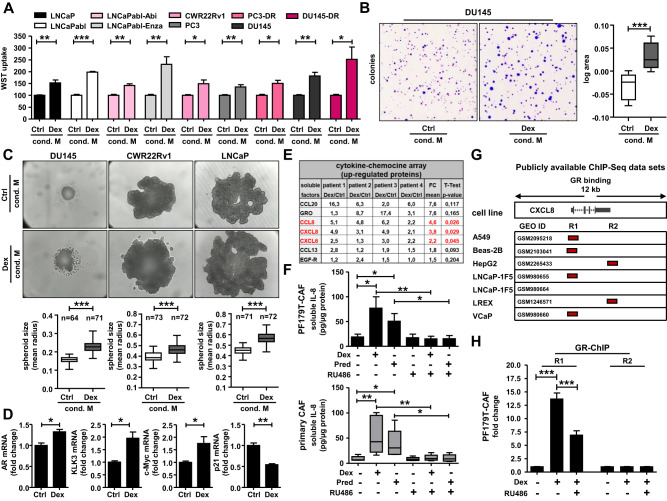
Fig. 4. Altered chemokine signaling results in accelerated cancer cell growth.
A Measurement of elevated cell proliferation/viability after 4 d incubation of several epithelial PCa cell lines, representing different tumor stages with collected conditioned stromal medium of PF179T-CAFs after 3 d control or 100 nM Dex treatment. Data represent mean + SEM from 3 independent experiments (unpaired t-test; *P < 0.05; **P < 0.01; ***P < 0.001). B Significantly increased DU145 colonies size after 12 d culture with collected conditioned stromal PF179T-CAF medium after previously 3 d control or 100 nM Dex treatment. Data represent the mean log area of 5616 measured colonies from 5 independent experiments (paired t-test; ***P < 0.001; Box Whisker Plot with min-max percentile). C Elevated 3D-spheroid growth of DU145, CWR22Rv1, and LNCaP cells after 8 d culture with collected conditioned stromal PF179T-CAF medium after previously 3 d control or 100 nM Dex treatment. Data represent mean area measured from 3 independent experiments (paired t-test; *P < 0.05; **P < 0.01; ***P < 0.001, Box Whisker Plot with min–max percentile). D Observed elevated AR, KLK3, and c-Myc, as well as decreased p21 mRNA expression after 8 d LNCaP spheroid growth with collected conditioned stromal PF179T-CAF medium after previously 3 d control or 100 nM Dex treatment. Data represent mean + SEM from 3 independent experiments (unpaired t-test; *P < 0.05; **P < 0.01). E Measurement of elevated soluble factors using commercially available cytokine and chemokine protein arrays after 3 d 100 nM Dex treatment using primary isolated CAFs from 4 PCa patients. Data represent mean fold change (paired t-test; marked in red, P < 0.05). F Altered IL-8 protein expression in PF179T-CAF cells and primary isolated CAFs after 3 d single/combination treatments with 100 nM Dex, 100 nM Pred, and 6 µM RU486, using specific IL-8-coupled magnetic Luminex® beads. Data represent mean + SEM from 3 independent experiments (one-way ANOVA and correction for multiple testing using Bonferroni’s comparison test; *P < 0.05; **P < 0.01). G Identification of 2 GR-binding sites (R1 and R2) near the CXCL8 gene in A549, Beas-2B, HepG2, LNCaP-1F5, LREX, and VCaP, cells, screening publicly available ChIP-Seq datasets. H GR-ChIP using PF179T-CAF cells after treatment with 100 nM Dex alone or with 6 µM RU486 for 16 h. Data represent mean + SEM from 3 independent experiments (one-way ANOVA and correction for multiple testing using Bonferroni’s comparison test; ***P < 0.001).