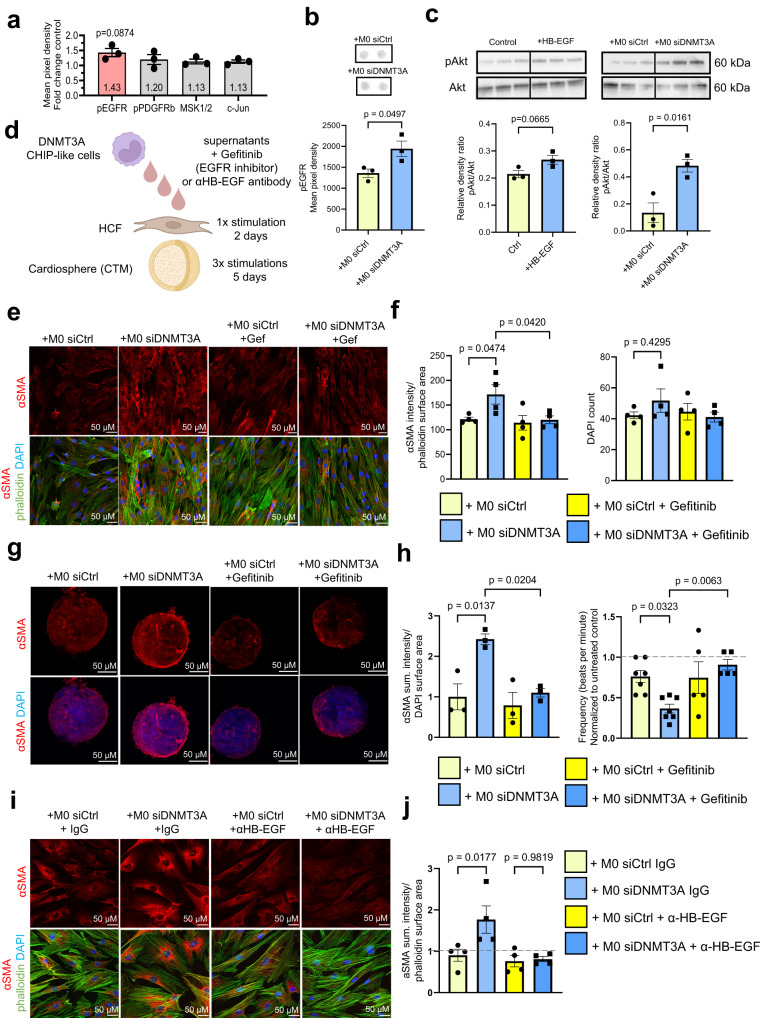
Fig. 8. Inhibition of HB-EGF-EGFR signaling reduces CHIP monocyte-mediated cardiac fibroblast activation.
a Analysis of phosphorylation of receptors and kinases in iHCF after incubation with supernatant from DNMT3A-silenced THP-1-derived cells for 30 min (n = 3 biologically independent experiments). Source data are provided as a Source Data file. b Quantification of EGFR phosphorylation (n = 3 biologically independent experiments) in iHCF after incubation with supernatant from DNMT3A-silenced THP-1-derived cells for 30 min. Source data are provided as a Source Data file. c Western blot for phospho-Akt (pAkt) and total Akt in iHCF stimulated with HB-EGF or with DNMT3A silenced monocytes (n = 3 biologically independent experiments). Source data are provided as a Source Data file. d Schematic representation of the experimental design used in (e–h). e Immunofluorescence analysis of HCF stimulated with DNMT3A silenced monocyte supernatants with or without gefitinib (5 μM) (n = 4 biologically independent experiments). f αSMA protein expression and cell numbers were determined by αSMA and DAPI staining in experiments shown in (e) (n = 4 biologically independent experiments). Source data are provided as a Source Data file. g Immunofluorescence analysis of cardiospheres stimulated with supernatant from DNMT3A silenced THP-1-cells with or without gefitinib (5 μM) (n = 3 biologically independent experiments). h αSMA protein expression and beating frequency of cardiospheres stimulated with supernatant from DNMT3A silenced THP-1-cells with or without gefitinib (n = 3 for αSMA protein expression in biologically independent experiments, beating frequency in n = 7 for both groups without gefitinib and n = 5 for both groups with gefitinib in biologically independent experiments) in experiments shown in (g). Source data are provided as a Source Data file. i Immunofluorescence analysis of HCF stimulated with supernatants from DNMT3A-silenced THP-1-cells with or without HB-EGF neutralizing antibody (50 μg/ml, 48 h) (n = 4 biologically independent experiments). j αSMA protein expression in experiments shown in (i) (n = 4 biologically independent experiments). Source data are provided as a Source Data file. Data are shown as mean ± SEM. a–c, f, h, j Normal distribution was assessed using the Shapiro–Wilk test. a-c, f, h, j Statistical analysis was performed using unpaired, two-sided Student’s t tests (b, c). t = 2.783, 4 degrees of freedom (b); t = 2.503, 4 degrees of freedom; t = 4.004, 4 degrees of freedom (c). Statistical analysis was performed using ordinary one-way ANOVA with Dunnett’s correction for pairwise comparisons for data with a Gaussian distribution (f); ordinary one-way ANOVA with post hoc Tukey tests for data with a Gaussian distribution (panel left) and a non-parametric Kruskal-Wallis test with Dunn’s multiple comparisons test for data without a Gaussian distribution (panel right) (h); ordinary one-way ANOVA with Šídák’s multiple comparisons test for data with a Gaussian distribution (j).