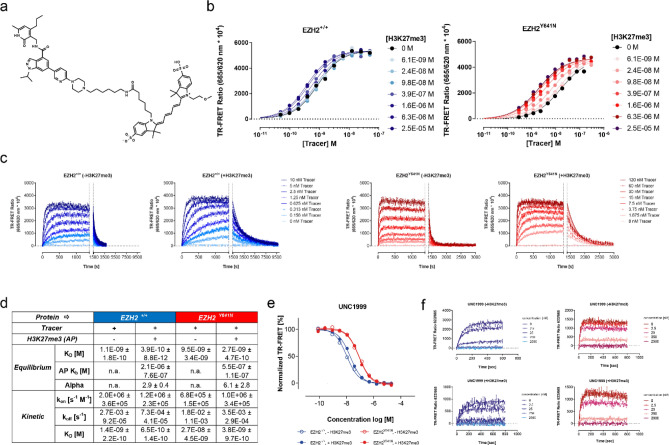
Figure 1.
Development of a fluorescent probe and homogeneous TR-FRET competition assays for the characterization of EZH2 inhibitors. (a) Chemical structure of the fluorescent probe synthesized for this study. (b) Equilibrium titrations of the probe on wild type (+/+) and mutant (Y641N) EZH2 performed in presence of increasing concentrations of H3K27me3 allosteric activator peptide (indicated on the right-hand side of the graphs). Fitting these curves to 1:1 single site- and allosteric binding models allowed calculating the KD and Kb values shown in panel d and the Supplementary Table S1. Here and along the entire paper, data for EZH2+/+ and EZH2Y641N are represented by blue and red dots and lines, respectively. (c) Kinetic titrations of the probe on EZH2+/+ and EZH2Y641N in the absence (left panel) or presence (right panel) of saturating concentrations of H3K27me3 activator peptide. Increasing concentrations of the probe (indicated on the right-hand side of each pair of graphs) were mixed with labeled enzyme and TR-FRET signals were recorded for 4 min. Equilibrium complexes were then disrupted by the addition of an excess amount of the unlabeled compound (marked with dotted vertical lines in the graph) and dissociation of the probe was followed until baseline fluorescence ratios were reached. Fitting of these kinetic traces to a global association and dissociation model led to the probe rate- and affinity constants shown in panel d, and the Supplementary Table S1. (d) Equilibrium and kinetic binding parameters of the fluorescent probe on EZH2+/+ and EZH2Y641N in the absence or presence of H3K27me3 activator peptide. (e) Exemplary normalized equilibrium curves of fluorescent probe displacement from wild type and mutant EZH2 by reference inhibitor UNC1999. Open and filled circles show competition in the absence and presence of saturating concentrations of the H3K27me3 peptide. Fitting these curves to a 4-parameter logistic model (solid lines), and conversion of the resulting IC50 values to Ki values with the Cheng-Prusoff equation, led to affinity parameters shown in the Supplementary Spreadsheet. (f) Representative kinetic probe competition assay (kPCA) traces for the same exemplary compound in the absence and presence of saturating concentrations of the H3K27me3 peptide. Fitting these curves to the Motulsky and Mahan model for competitive binding kinetics (solid lines) yielded the kinetic parameters shown in the Supplementary Spreadsheet.