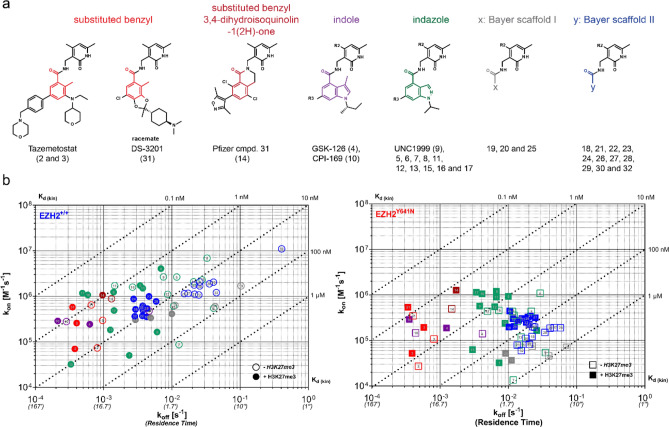
Figure 2.
Kinetics of EZH2 binding by pyridone-based active site inhibitors. (a) Structural features of the EZH2 inhibitor set investigated in this study. All compounds share a common pyridone motif in the north side of the molecule and are differentiated by distinct chemical cores highlighted in different colors. For two of the Bayer-proprietary chemical series, distinctive colors were only applied to label the linkage between the pyridone and the undisclosed heterocyclic scaffolds. Whenever relevant (and feasible), this color code was subsequently used to label compounds throughout the entire paper. (b) Rate plots with isoaffinity diagonals (RAPID) representing the binding to wild type and mutant EZH2 of all compounds investigated in this study. Compounds are colored and numbered as in panel a for visualization of structure kinetic relationships (SKR). Open or filled symbols represent data generated in the absence or presence of H3K27me3 peptide.