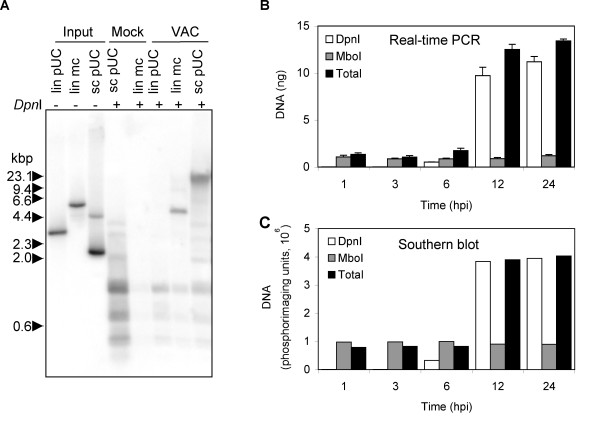
Figure 1.
Replication of transfected DNA in VAC-infected cells. (A) Southern blot of replicated circular plasmid and linear minichromosome. B-SC-1 cells were infected with VAC and 1 h later transfected with equal molar amounts (20 fmol) of super coiled pUC13 (sc pUC), pUC13 linearized by digestion with EcoRI (lin pUC), linear minichromosome containing pUC13 and 1.3 kbp viral telomeric sequences (lin mc). As a control, cells were mock infected and transfected with 20 fmol of linear minichromosome or 10 times that amount (200 fmol) of super coiled pUC13. At 24 h after infection or mock infection, cells were collected and total DNA extracted. Total DNA (2 μg) was digested with DpnI subjected to agarose gel electrophoresis and analyzed by Southern blot hybridization using a 32P-labeled pUC13 probe. Samples (0.5 fmol of lin pUC, 0.5 fmol of lin mc, 1 fmol sc pUC) of the DNA used for transfections (input DNA) were also analyzed. The positions of marker DNA (kbp) are shown on the left. (B) Real-time PCR of replicated plasmid. BS-C-1 cells were transfected with the plasmid p716 at 24 h prior to infection with VAC. At indicated hours post infection (hpi), cells were harvested and total DNA extracted. DNA was untreated or treated with DpnI or MboI and analyzed by real-time PCR using primers specific to plasmid DNA. (C) Quantification of Southern blot. DNA described in panel (B) was digested with EcoRI prior to MboI or DpnI treatment. The digested DNA samples were subjected to gel electrophoresis, transferred to a Nylon membrane, hybridized to a 32P-labeled p716 probe, and the radioactivity quantified with a phosphoImager.