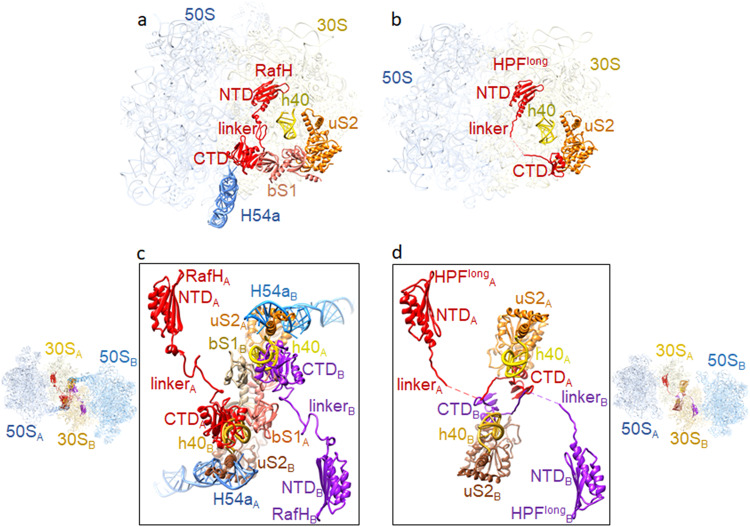
Fig. 6. Comparison of RafH binding in 70S ribosome with HPFlong binding in 100S ribosome.
a RafH, bS1, uS2, and h40 of 16S rRNA and H54a of 23S rRNA are shown with LSU and SSU in the 95% transparent background. b The corresponding position of HPFlong, uS2, and h40 of 16S rRNA in one of the ribosomes of the Staphylococcus aureus 100S structure (PDB ID; 5NGM) is shown with LSU and SSU in the 95% transparent background. c Two 70S ribosome RafH complex structures docked into the corresponding positions in S. aureus 100S dimer structure (PDB ID; 6FXC) and RafH CTD interacting components are shown in 80% transparent background on the left side and magnified view with a white background are shown in the box on the right side. One 70S ribosome is labeled as A, and the other 70S ribosome is labeled as B. d The HPFlong interacting components uS2 and h40 of 16S rRNA in S. aureus 100S ribosome dimer interface (PDB ID; 6FXC) are shown on the right side with 30S and 50S in 80% transparent background, and a magnified view with white background is shown in the box on the left side. Similar to (c), one 70S ribosome is labeled as A, and the other 70S ribosome is labeled as B. A multiple sequence alignment among HPFs is shown in Supplementary Fig. 9.