FIGURE 4.
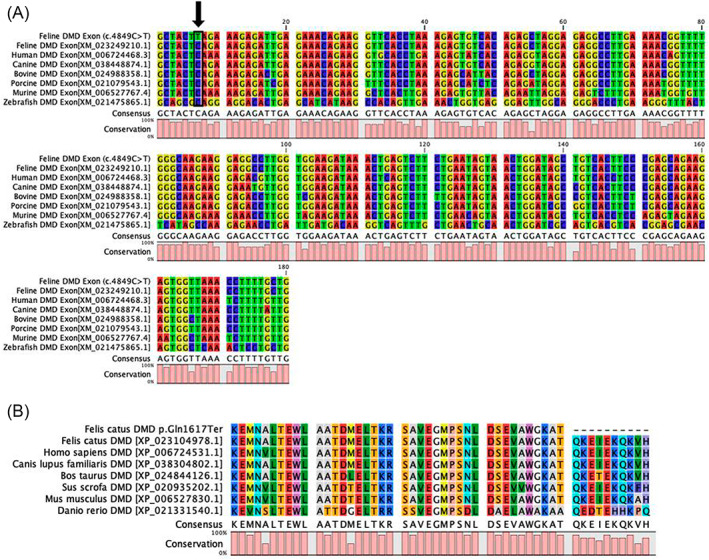
Multispecies alignment of Felis catus DMD exon 35. The alignments indicate the position of the DMD stop gain LoF variant (XM_023249210.1, ENSFCAT00000068370:c.4849C>T; p.Gln1617Ter) at chromosome position X:28201541 Felis_catus_9.0 at the beginning of exon 35. (A) Nucleotide alignment for diverse species (DMD affected feline case, normal feline, human, canine, bovine, porcine, murine, and zebrafish) indicating the 100% conservation of the nucleotide and the high conservation of the exon 35, lower histogram. Nucleotides are represented by the different colors. (B) Protein alignments for the same species depicting the termination in exon 35 for the affected cat. The alanine that is 2 amino acids upstream of the amber termination codon is the 5′ start of exon 35. The mammals have nearly 100% amino acid conservation within this region.
