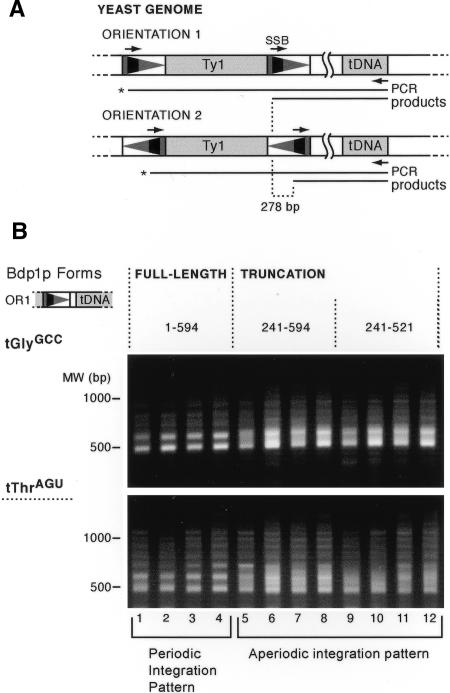
Figure 1.
PCR assay and pattern of Ty1 integration in bdp1 N-terminal deletion mutants. (A) Schematic of the PCR assay used in this work. Galactose-induction of a plasmid-borne transposon pVIT41 containing a unique sequence marker (“SSB,” narrow black rectangle) in the LTR (triangle) results in integration into the yeast genome. PCR using the SSB primer and a primer complementary to the tDNA results in amplification of a population of insertion events upstream of genes within the same tDNA family. Use of a primer complementary to a unique region downstream of a specific tDNA amplifies insertions upstream of a single tDNA target. Insertions in both orientations can be amplified using either the SSB primer or its reverse complement. Because of the asymmetric position of the SSB in the LTR, PCR products generated with the orientation 1 (OR1) primer will be 278 bp larger than those using the reverse complement, orientation 2 (OR2). (B) Removal of the first 240 residues of Bdp1p results in altered patterns of Ty1 insertions upstream of tGlyGCC (16 copies in the yeast genome) and tThrAGU genes (11 copies). Strains expressing either the wild-type Bdp1p (1-594) or the truncation alleles (241-594 and 241-521) were transformed with pVIT41, and four independent transformants were induced for transposition. Insertions upstream of tGlyGCC (top) and tThrAGU (bottom) were analyzed by PCR using the orientation 1 primer.