SUMMARY
The receptor-binding domain (RBD) of influenza virus hemagglutinin (HA) elicits potently neutralizing yet mostly strain-specific antibodies. Here, we evaluate the ability of several immunofocusing techniques to enhance the functional breadth of vaccine-elicited immune responses against the HA RBD. We present a series of “trihead” nanoparticle immunogens that display native-like closed trimeric RBDs from the HAs of several H1N1 influenza viruses. The series includes hyperglycosylated and hypervariable variants that incorporate natural and designed sequence diversity at key positions in the receptor-binding site periphery. Nanoparticle immunogens displaying triheads or hyperglycosylated triheads elicit higher hemagglutination inhibition (HAI) and neutralizing activity than the corresponding immunogens lacking either trimer-stabilizing mutations or hyperglycosylation. By contrast, mosaic nanoparticle display and antigen hypervariation do not significantly alter the magnitude or breadth of vaccine-elicited antibodies. Our results yield important insights into antibody responses against the RBD and the ability of several structure-based immunofocusing techniques to influence vaccine-elicited antibody responses.
Graphical Abstract
In brief
Dosey et al. have engineered a series of influenza hemagglutinin (HA) head-based nanoparticles with increasing amounts of immune focusing onto the conserved receptor-binding site. They show the effect that each of these focusing strategies has on the potency, breadth, and direction of the elicited immune response.
INTRODUCTION
Influenza viruses currently persist as a major public health threat due to their high evolutionary rate, which gives rise to frequent antigenic drift among circulating strains.1,2 This strain divergence is due in large part to diversity in the surface glycoprotein hemagglutinin (HA) as a result of immune pressure.3,4 Despite its variation among strains, HA characterization has revealed two functionally conserved sites: the receptor-binding site (RBS) in the head domain, which mediates host cell entry by binding to sialic acid on host glycoproteins, and a conserved antigenic region in the HA stem that is involved in host membrane fusion.5 The identification of several broadly neutralizing antibodies (bNAbs) against these sites has made them central targets in vaccine design efforts.6–13 The HA head is immuno-dominant, and antibodies that bind near the RBS typically exhibit potent neutralization by blocking receptor binding.5,14,15 However, high levels of antigenic variation in the head domain allow influenza viruses to evade head-directed immunity through antigenic drift.16 Thus, bNAbs targeting the conserved RBS itself are rarely elicited by infection or vaccination, and response breadth is often limited by frequent residue mutations in the RBS periphery.17 By contrast, antibodies targeting the central stem epitope tend to react more broadly to HAs from different influenza viruses, but these antibodies are not always neutralizing, and they are difficult to elicit due to the immune subdominant nature of the stem. Recently, broadly protective antibodies against additional antigenic sites on HA have been discovered, including the stem anchor epitope18,19 and the trimer interface in the head domain.20–22 The broad binding of trimer interface-directed antibodies against divergent HA subtypes makes this site an intriguing vaccine target. Although they lack neutralizing activity in vitro, initial reports indicate that trimer-interface-directed antibodies can be protective in animal models.20,21
In addition to amino acid substitutions, influenza viruses use glycosylation of their surface proteins as a mechanism of immune evasion. Analysis of HA sequences obtained over the past century has revealed that the HA head has acquired more glycans over time, and this has been shown to both prevent antibody binding and to lower the number of mutations required for immune evasion.23,24 These observations have inspired the use of glycan engineering in vaccine design as a means to direct immune responses to target epitopes. Previous studies have generated hyperglycosylated immunogens by introducing new N-linked glycosylation motifs in the immune-dominant head domain, resulting in the diversion of antibody responses onto conserved, subdominant epitopes in either the stem domain25 or the trimer interface.26 A reduction in non-neutralizing trimer interface responses was seen in a separate study that combined hyperglycosylation and disulfide bond engineering in full-length HA ectodomains, but this did not increase the elicitation of broadly reactive, RBS-directed antibodies.27
The utilization of variable sequences within vaccines as a means to focus immune responses onto conserved epitopes has been implemented in various formats. One approach threaded divergent HA sequences onto H3 or B HA antigens and then employed those varying immunogens in a sequential vaccination regimen, which resulted in protection from vaccine-mismatched challenge.28,29 Beyond heterologous prime-boost strategies, another strategy is the co-display of several variants of a given antigen on nanoparticle scaffolds, an approach known as “mosaic nanoparticle display.” In the first application of this strategy, the assembly of up to eight different HA receptor-binding domains (RBDs) on the same mosaic ferritin nanoparticle resulted in higher levels of cross-reactive B cells and significantly better neutralization breadth against a large panel of H1 viruses compared to the same RBD antigens presented in either a heterologous prime-boost regimen or as a cocktail of homotypic (i.e., single-strain) nanoparticles.30 Furthermore, a recent study applying this mosaic approach to the display of trimeric HA ectodomains elicited broadly protective immune responses that were subtly yet consistently superior to cocktails of homotypic particles,31 although another study using a different display approach failed to observe a similar effect.32 Mechanistically, it has been proposed that B cell receptors (BCRs) targeting conserved epitopes on mosaic nanoparticles have an avidity advantage compared to BCRs directed against variable epitopes and that this allows for increased antigen binding and thus B cell activation.30 To date, all reports of mosaic nanoparticle immunogens have tested co-display of antigens with wild-type sequences from various virus isolates; the use of synthetic or designed antigenic variation in mosaic nanoparticles remains unexplored.
Here, we evaluated the ability of several known and novel immunofocusing techniques—conformational stabilization, hyperglycosylation, mosaic nanoparticle display, and the design of synthetic “hypervariable” antigens—to focus antibody responses on the conserved RBS of the HA head. We found that combining conformational stabilization, hyperglycosylation, and mosaic nanoparticle display substantially altered the epitopes targeted by vaccine-elicited antibodies to elicit potent vaccine-matched and -mismatched responses, while additional co-display of hypervariable antigens did not further alter the potency or epitope specificity of the serum antibody response.
RESULTS
Design and immunogenicity of hyperglycosylated trihead nanoparticle immunogens
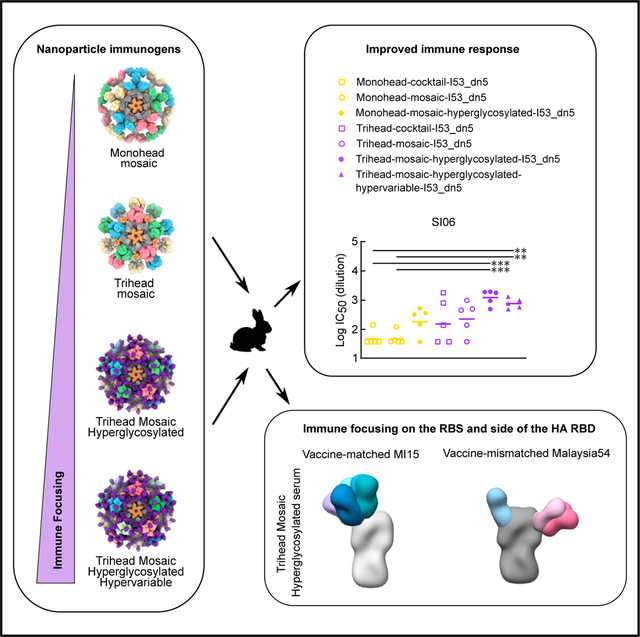
In our accompanying article in this issue of Cell Reports, we describe the design of a “trihead” antigen in which the RBD of H1 A/New Caledonia/20/1999 (NC99) was stabilized in a native-like trimeric state via hydrophobic mutations at the trimer interface and a rigid fusion to the trimeric component of the I53_dn5 nanoparticle.33 We also show that this stabilized immunogen, which we refer to here as TH-NC99 (Figure 1A), elicits potent neutralization and hemagglutination inhibition (HAI) activity in immunized mice. To maximize focus of immune responses elicited by this immunogen to the conserved RBS, three and five additional N-linked glycans were engineered into epitopes distant from the RBS to create the hyperglycosylated NC99 triheads TH-NC99–7gly and TH-NC99–9gly, respectively (Figure 1A). The new glycans were designed using the Rosetta modeling suite, both to guide sequon design and model glycan structure.34,35 TH-NC99 constructs with individual glycan additions were first evaluated for their secretion from HEK293F cells, and those that maintained expression were then combined. We also generated “monohead” antigens lacking the trimer-interface-stabilizing mutations and rigidifying disulfide bond, which form monomers with exposed trimer interfaces,33 bearing the four wild-type (MH-NC99) and five additional (MH-NC99–9gly) N-linked glycans. Amino acid sequences for all novel proteins used in this study can be found in Table S1. The monohead and trihead antigens were secreted from HEK293F cells as genetic fusions to the I53_dn5B trimer and purified via immobilized metal affinity chromatography (IMAC) and size-exclusion chromatography (SEC) (Figure S1A). SDS-PAGE revealed slower migration for the hyperglycosylated monohead and trihead sub-units compared to their wild-type counterparts (Figures 1B and S1B). Treatment of the trimeric components with PNGaseF resulted in a large decrease in apparent molecular weight by SDS-PAGE and uniform migration of all constructs. Biolayer interferometry (BLI) using the anti-RBS bNAb C056 showed similar binding profiles for all five components, indicating that the hyperglycosylated antigens maintain RBS antigenicity (Figure 1C). By contrast, BLI using the anti-trimer interface monoclonal antibody (mAb) FluA-2021 showed high binding to monohead components but minimal binding to all trihead components, confirming stable closure of the trimer interface in the trihead antigens. Lastly, BLI using the anti-lateral patch mAb Ab664936 showed nearly full binding to TH-NC99–7gly but greatly diminished binding to TH-NC99–9gly, likely due to the additional glycan at position 167 in TH-NC99–9gly that is in the center of the Ab6649 epitope. MH-NC99–9gly showed moderate Ab6649 binding, possibly due to less consistent glycosylation at that position than in TH-NC99–9gly.
Figure 1. Design and immunogenicity of hyperglycosylated NC99 trihead nanoparticle immunogens.
(A) Model structures and gene diagrams for wild-type and hyperglycosylated NC99 triheads with wild-type glycans in light purple and glycan knockins in dark purple. NC99 HA numbering is in blue, and trihead model numbering is in black.
(B) Reducing SDS-PAGE of wild-type and hyperglycosylated NC99 monoheads and triheads without and with PNGaseF digestion.
(C) BLI of wild-type and hyperglycosylated NC99 monoheads and triheads against C05, FluA-20, and Ab6649.
(D) nsEM micrographs of hyperglycosylated NC99 monohead and trihead I53_dn5 nanoparticles. Scale bars = 100 nm.
(E) Schematic illustrating mouse study timeline, immunizations, and serology timepoint.
(F) Week 10 NC99-foldon trimer ELISA titers plotted as the reciprocal EC50 titer, hemagglutination inhibition (HAI) titers, and the ratio of HAI/reciprocal EC50 titers of hyperglycosylated NC99 monohead and trihead nanoparticles in BALB/c mice. Each symbol represents an individual animal, and the geometric mean of each group is indicated by the bar (n = 5 mice/group).
Statistical significance was determined using one-way ANOVA with Tukey’s multiple comparisons test; *p < 0.05; **p < 0.01.
To quantify glycan occupancy at each site, all trihead components were analyzed using peptide mass spectrometry.37,38 Glycan occupancy in TH-NC99 was high across all sites with the exception of N163, which was only about half occupied (45.1%; Table S2). The wild-type sites in TH-NC99–7gly showed similar occupancy, including half occupancy at N163 (44.3%). The engineered glycosylation sites in TH-NC99–7gly had high occupancy at N125b, about half occupancy at N81 (46.2%), and low occupancy at N171 (20.1%). The additional site at N125b was not observed as an isolated glycopeptide and could only be indirectly quantified by the peptide spanning the additional glycan site at N129. For TH-NC99–9gly, the additional N77 glycosylation site was only observed in a glycopeptide that also comprised N81 and showed either single (63%) or double (37%) occupancy. The last two sites in TH-NC99–9gly were only observed on a long glycopeptide containing three sequons. Based on the occupancy levels measured, it is likely that N167 is highly occupied (>90%) while N171 is less than 15% occupied, as this would be consistent with the occupancy levels observed for these sites in TH-NC99–7gly.
The monohead and trihead components were then combined with purified I53_dn5A pentamer in vitro at a 1:1 M ratio to form icosahedral I53_dn5 nanoparticles displaying either 60 monohead monomers or 20 trihead trimers. Purification by SEC (Figure S1C) yielded pure, monodisperse preparations of nanoparticles according to SDS-PAGE, negative-stain electron microscopy (nsEM), and dynamic light scattering (DLS), with a small amount of residual unassembled component detectable by nsEM (Figures 1D, S1D, and S1E). Efficient assembly was observed for all trihead nanoparticles, as little residual component was observed during SEC purification (Figure S1C; peak at 17 mL). By contrast, substantial residual component was observed during SEC of the monohead nanoparticle assemblies, indicating less efficient assembly.
We evaluated the hyperglycosylated trihead nanoparticle immunogens in an initial immunogenicity study in mice. BALB/c mice were immunized with 1.5 μg nanoparticle immunogen formulated with AddaVax at weeks 0, 4, and 8 (Figure 1E). Similar binding titers against the I53_dn5 nanoparticle scaffold were seen across all groups (Figures S1F and S1G). Strain-matched NC99 HA-binding antibody titers from serum collected at week 10 showed reduced binding in all hyperglycosylated groups compared to their wild-type counterparts (Figure 1F). Conversely, NC99 HAI titers were highest in the TH-NC99–9gly group and lowest in the MH-NC99–9gly group. Plotting the ratio of HAI/HA-binding titers revealed a trend toward a stepwise increase with increasing glycosylation in the trihead groups, suggesting a higher proportion of on-target receptor-blocking antibodies. Only the MH-NC99–9gly sera competed with FluA-20 binding in competition ELISAs, yet these sera showed the least amount of competition with C05 (Figure S1H). These results suggest that hyperglycosylation refocused vaccine-elicited antibodies onto receptor-blocking epitopes in the case of the trihead immunogens and onto the trimer interface in the case of the monohead immunogens.
Design of hyperglycosylated trihead antigens from additional H1 HAs
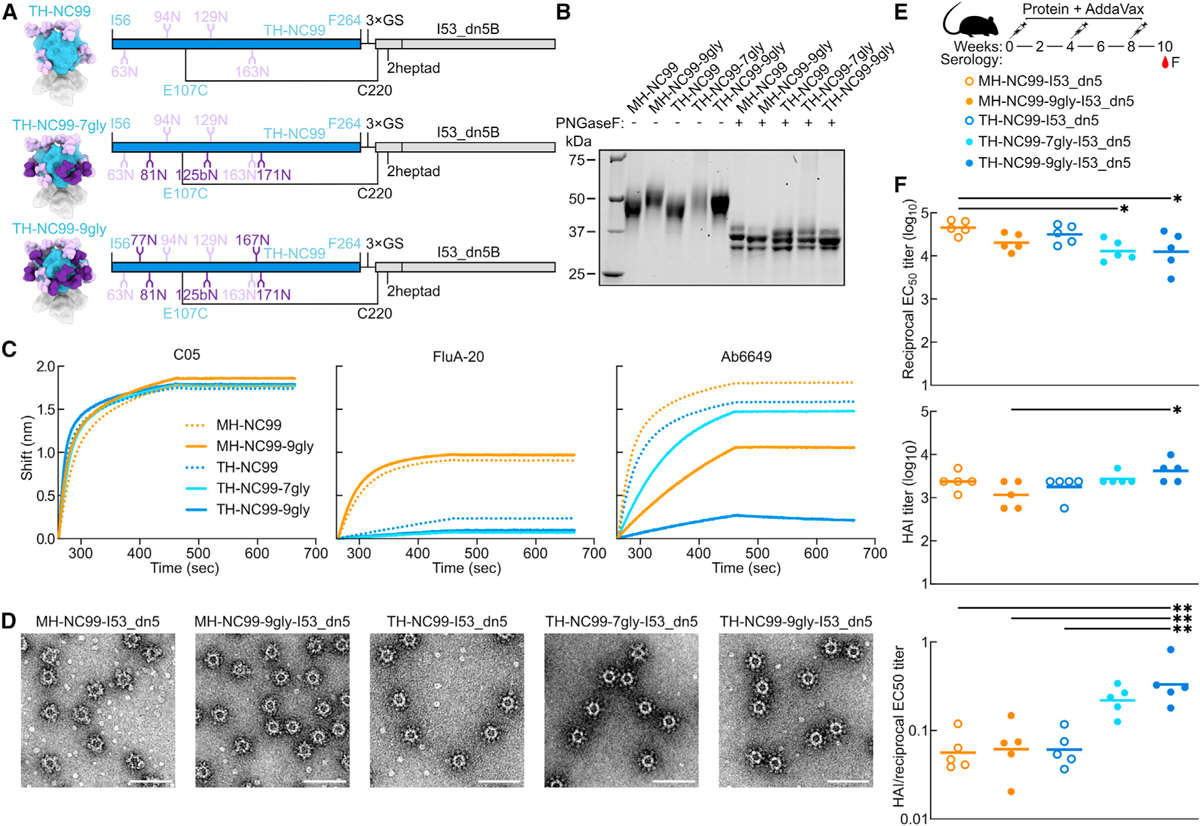
We and others have recently reported that mosaic nanoparticle immunogens, which co-display multiple antigenic variants on the same nanoparticle surface, can induce broadly protective responses against related viruses by eliciting antibodies that target conserved epitopes.30,31,39–43 To enable mosaic trihead display as a potential route to enhancing breadth among H1 strains, we adapted the trihead design strategy to three other divergent H1s with unique antigenic properties: A/South Carolina/1/1918 (TH-SC18), A/Puerto Rico/8/1934 (TH-PR34), and A/Michigan/45/2015 (TH-MI15). We again made corresponding monohead antigens for comparison. These antigens were all connected to the I53_dn5B trimer using one heptad repeat of the GCN4-based coiled coil, as this rigid linker length was found to yield optimal cross-reactive antibody responses in mice.33 The same disulfide bond in TH-NC99 between the base of the trihead and the coiled-coil linker was used, as well as similar stabilizing mutations at the trimer interface, although the amino acids used at positions 203 and 205 differed among strains (Figure 2A; Table S1). Glycan knockin mutations were included in final designs for TH-PR34 at position 63 and for MH-SC18 and TH-SC18 at position 125b, which dramatically enhanced secretion and stability. Additional resurfacing mutations P26S and V84E in TH-PR34, as well as A198E in both TH-SC18 and TH-MI15, were also key to enhancing secretion. The Y98F mutation was also included in all trihead and monohead constructs to knock out sialic acid binding and promote secretion.44,45
Figure 2. Design of hyperglycosylated trihead antigens from additional H1 HAs.
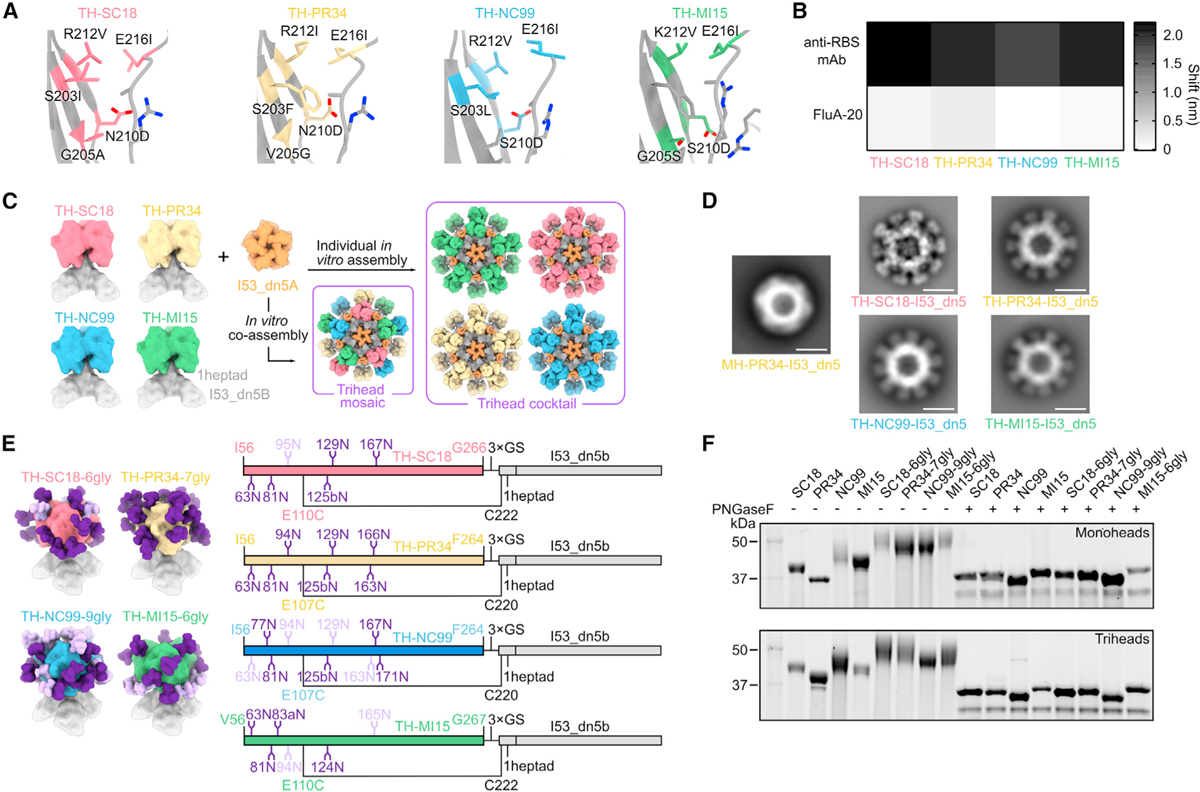
(A) Diagram of RBD trimer interfaces for TH-SC18, TH-PR34, TH-NC99, and TH-MI15, where mutated residues are colored and labeled.
(B) BLI of trihead components against RBS-directed mAbs (5J8, anti-PR34, and C05) and FluA-20.
(C) Schematic of TH-SC18, TH-PR34, TH-NC99, and TH-MI15 constructs and their in vitro assembly into mosaic or cocktail I53_dn5 nanoparticles.
(D) nsEM 2D class averages of MH-PR34-I53_dn5 and trihead I53_dn5 nanoparticles. Scale bars = 25 nm.
(E) Model structures and gene diagrams for hyperglycosylated triheads with wild-type glycans in light purple and glycan knockins in dark purple. Strain-specific H1 HA numbering is in respective HA strain color, and trihead model numbering is in black.
(F) Reducing SDS-PAGE of wild-type and hyperglycosylated monoheads and triheads without and with PNGaseF digestion.
All four triheads maintained binding to RBS-directed antibodies, with minimal FluA-20 binding by BLI, indicating trihead closure (Figures 2B and S2A). By contrast, monohead versions of each strain all showed high binding to both RBS antibodies and FluA-20. The trihead and monohead components were purified using SEC (Figures S2B and S2C) prior to in vitro assembly into I53_dn5 nanoparticles. We prepared a cocktail of nanoparticles by mixing together the four individually assembled trihead nanoparticles, as well as mosaic nanoparticles in which the four trihead components were mixed together prior to addition of I53_dn5A pentamer (Figure 2C). We have shown previously that the latter approach allows precise control over the overall amount of each antigen in the preparation, although the distribution of antigens on each nanoparticle is stochastic.31 All nanoparticles were then purified using SEC, and their purity and monodispersity were verified by SDS-PAGE, DLS, and nsEM (Figures S2D–S2G). All four monovalent trihead nanoparticles exhibited individually resolved trihead densities on the nanoparticle exteriors in nsEM averages, while the MH-PR34-I53_dn5 nanoparticle lacked visible antigen density due to the flexibility of the monoheads (Figure 2D). Taken together, the BLI, SEC, and nsEM averages indicate the formation of closed, relatively rigid trihead nanoparticle immunogens for all four strains.
As with TH-NC99–9gly, engineered glycans were used to mask epitopes outside the RBS in each H1 trihead. Sites for glycan introduction were chosen by ensuring that Rosetta-modeled N-linked glycans were sterically accommodated34 and by avoiding mutations to any buried residues. Individual glycans added to TH-SC18 were tested for their effects on trihead secretion before combination into one hyperglycosylated construct. These data were then used to guide hyperglycosylation of TH-PR34 and TH-MI15. TH-SC18–6gly has one wild-type and five engineered glycans, TH-PR34–7gly has zero wild-type and seven engineered glycans, and TH-MI15–6gly has two wild-type and five engineered glycans (Figure 2E; Table S1). All hyperglycosylated trihead components showed apparent increases in molecular weight by SDS-PAGE compared to their wild-type counterparts, while treatment with PNGaseF resulted in nearly uniform migration of all constructs at a lower apparent molecular weight (Figures 2F and S2C). All hyperglycosylated trihead components maintained the desired antigenic profile of high anti-RBS antibody binding with minimal FluA-20 binding and had circular dichroism (CD) spectra that closely matched those of the corresponding non-hyperglycosylated triheads, indicating they retained their native structure (Figures S3A–S3C). The four hyperglycosylated trihead components were combined in an equimolar mixture and then co-assembled in vitro with the I53_dn5A pentamer to form a hyperglycosylated trihead mosaic nanoparticle that was purified by SEC (Figure S2D). SDS-PAGE, DLS, and nsEM of the purified assembly revealed monodisperse nanoparticles of the expected size and morphology (Figures S2E–S2G).
We compared the stabilities of the hyperglycosylated monohead and trihead components using hydrogen-deuterium exchange mass spectrometry (HDX-MS) and nanodifferential scanning fluorimetry (nanoDSF). Deuterium uptake profiles across MH-NC99–9gly and all four hyperglycosylated triheads were consistent with the antigens adopting similar conformations, albeit with some local differences (Figure S4A). For example, HDX-MS analysis of RBS peptides showed that TH-SC18–6gly has greater dynamics at the 190 helix and 220 loop relative to the other three strains of hyperglycosylated triheads (Figures S4B and S4C). Similarly, trimer interface peptides lying in the 220 loop and the 200 loop in the TH-SC18–6gly were also less ordered, especially compared to TH-NC99–9gly and TH-PR34–7gly, which were the most ordered in these regions. Comparing MH-NC99–9gly and TH-NC99–9gly showed that the monohead antigen displayed substantially higher exchange than the trihead at the trimer interface in the 220 loop, as well as slightly elevated dynamics in other regions. Thermal denaturation monitored by SYPRO Orange fluorescence showed similar trends in stability, where TH-SC18–6gly had the lowest melting temperature (Tm) and TH-NC99–9gly had the highest Tm among the triheads (Figure S4D). Additionally, three out of four hyperglycosylated monoheads had Tms ≥4°C lower than their trihead counterparts. Together, these analyses indicate that the trihead antigens are locally and globally more stable than monomeric RBDs in addition to being rigidly linked to the I53_dn5 nanoparticle scaffold.
Design and characterization of hypervariable trihead immunogens
As a further test of the hypothesis that mosaic nanoparticle display focuses antibody responses on conserved epitopes, we designed a hypervariable antigen library featuring mutations within the RBS periphery. This is a particularly variable region in HAs from different influenza virus strains, and mutations in this region are a central driver of antigenic drift (Figure 3A).3 We reasoned that co-display of a library of trihead variants with mutations in the RBS periphery may elicit fewer strain-specific antibodies in favor of responses targeting the conserved RBS. We constructed our library to recapitulate this variability by introducing naturally occurring mutations or those identified by deep mutational scanning into key hypervariable positions (Figure 3A).46,47 We made four variants of each hyperglycosylated trihead, each comprising a unique combination of 2–10 amino acid mutations in the RBS periphery (Figure 3B; Table S3). Binding studies using BLI showed that the variants had distinct antigenic profiles as intended, with some mutations leading to a complete loss of binding to particular anti-RBS mAbs (Figures 3C and S3A). However, all RBS variants maintained minimal FluA-20 binding, exhibited the expected SEC elution profiles and apparent molecular weights on SDS-PAGE, and had similar CD spectra showing a mixture of alpha helix and beta sheet, indicating that they formed well-folded, closed triheads (Figures 3C, S3C, S5A, and S5B). The four base hyperglycosylated triheads and all of their variants were then pooled and co-assembled with I53_dn5A pentamer to generate a hyperglycosylated, hypervariable nanoparticle containing 20 unique trihead antigens (Figure 3D). This hypervariable trihead nanoparticle was purified by SEC and was monodispersed by SDS-PAGE, DLS, and nsEM (Figures S2D–S2G). Co-display of all trihead mosaic nanoparticles (TH-mosaic-I53_dn5, TH-hyperglycosylated-mosaic-I53_dn5, and TH-hypervariable-hyperglycosylated-mosaic-I53_dn5) was confirmed using sandwich BLI by comparison to the TH-cocktail-I53_dn5 immunogen (Figure S5C). The NC99-reactive mAb C05 was first loaded onto AR2G biosensors, followed by nanoparticle loading and sequential binding to a PR34-specific mAb (Sino Biological) and 5J8, which binds both MI15 and SC18. Although all nanoparticles bound to the immobilized C05 mAb, only the three mosaic nanoparticles showed subsequent binding to the PR34-specific mAb and 5J8. These data indicate that the mosaic nanoparticles co-display trihead antigens that bind all three antibodies and that there is no detectable subunit exchange in the cocktail nanoparticle preparation. We note that the lower amount of hypervariable trihead nanoparticle loading and subsequent antibody binding is consistent with the individual hypervariable trihead component BLI, which demonstrated that some mutations within the RBS periphery abrogated specific mAb binding, particularly for the anti-PR34 mAb (Figures 3C and S3A).
Figure 3. Design and characterization of hypervariable trihead immunogens.
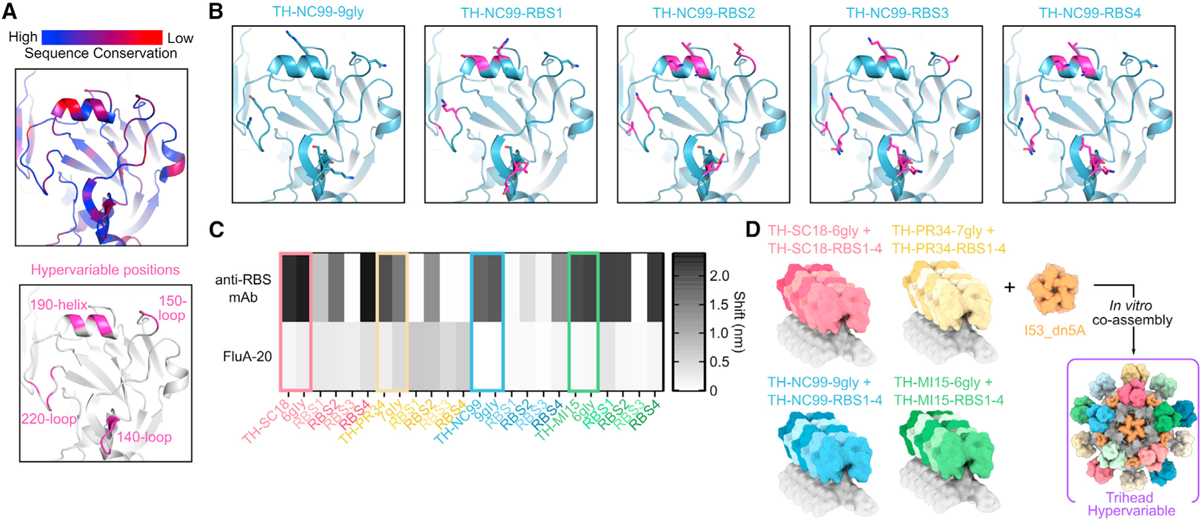
(A) Sequence conservation among 643 unique H1 sequences (top) and positions mutated in hypervariable library as dark pink (bottom) modeled on the NC99 HA structure (PDB: 7SCN).
(B) TH-NC99–9gly wild-type and hypervariable variants modeled onto the NC99 HA structure (PDB: 7SCN), with all positions mutated in the library shown as sticks, wild-type residues in blue, and mutated residues in magenta.
(C) BLI of triheads and hyperglycosylated triheads, with colored squares around these constructs, and trihead RBS variant components against RBS-directed mAbs (5J8, anti-PR34, and C05) and FluA-20.
(D) Schematic of hypervariable trihead components and assembly into an I53_dn5 nanoparticle.
Vaccine-elicited antibody responses in rabbits immunized with monohead and trihead nanoparticles
We then evaluated our series of monohead and trihead nanoparticle immunogens in an immunogenicity study in New Zealand white rabbits. We chose rabbits because their long CDRH3 repertoire may facilitate the elicitation of antibodies that can penetrate into the RBS6,7,48 and also because they permit the collection of sufficient serum to conduct a number of distinct serological analyses. Rabbits were immunized at weeks 0, 4, and 20 with 25 μg immunogen formulated with AddaVax (Figure 4A). Negligible binding titers at week 0 against a vaccine-matched MI15 HA-foldon trimer showed that there were no pre-existing anti-HA antibodies in these animals (Figures S5D and S5E). There were very similar binding titers in week 22 serum to the I53_dn5 nanoparticle scaffold across all groups (Figure S5F). Evaluation of vaccine-matched (NC99 and MI15) serum antibody binding, HAI, and microneutralization using sera obtained at weeks 6 and 22 revealed several differences between the groups. First, the cocktail and mosaic monohead groups consistently had lower binding, HAI, and neutralization titers than all other groups at week 6, although the differences were not always statistically significant (Figures 4B–4D). Interestingly, the hyperglycosylated monohead mosaic elicited antibody responses that were significantly higher than the other monohead groups and comparable to the trihead immunogens. At week 22, the monohead cocktail group had significantly lower NC99-neutralizing activity than all other groups, as well as NC99 HAI titers that trended lower (Figures 4E and S5G). By contrast, the monohead and trihead mosaic groups had significantly lower MI15-neutralizing activity than most of the other groups (Figure 4E). Taken together, the data show that the monohead cocktail and mosaic nanoparticle immunogens were generally less immunogenic than the other groups, while the hyperglycosylated and hypervariable nanoparticles consistently elicited potent vaccine-matched responses.
Figure 4. Vaccine-elicited antibody responses in rabbits immunized with monohead and trihead nanoparticles.
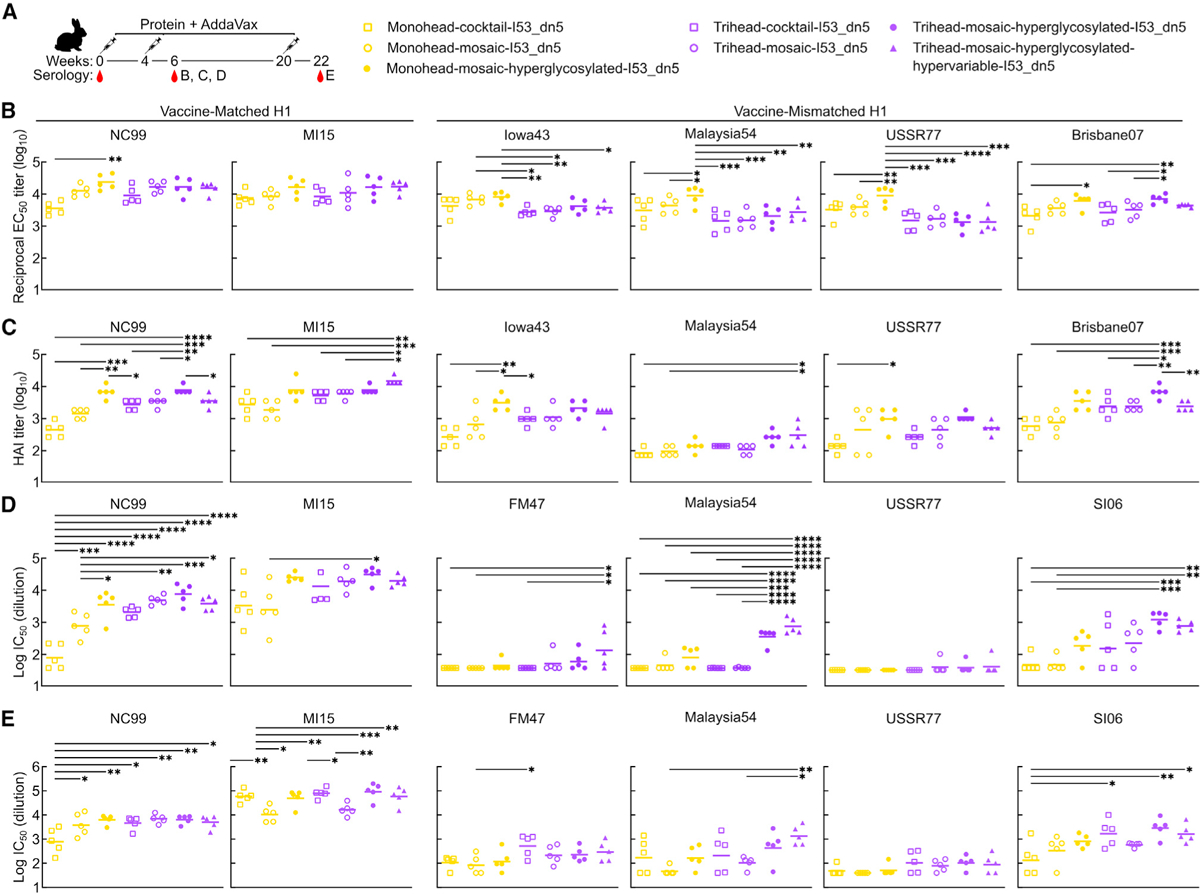
(A) Hypervariable trihead nanoparticle rabbit immunization schedule and groups.
(B–D) ELISA binding titers(B), HAI titers (C), and microneutralization titers (D) in immune sera at week 6.
(E) Microneutralization titers at week 22.
Each symbol represents an individual animal, and the geometric mean of each group is indicated by the bar (n = 5 rabbits/group). Statistical significance was determined using one-way ANOVA with Tukey’s multiple comparisons test; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Across four vaccine-mismatched H1 strains (see Table S1), the hyperglycosylated monohead group had the highest binding titers of all groups, both at weeks 6 and 22 (Figures 4B and S5H). The hyperglycosylated monohead also induced relatively high levels of HAI; however, it elicited lower neutralizing activity than the hyperglycosylated trihead groups for all mismatched H1 strains at both weeks 6 and 22, although this was not always significant (Figures 4C–4E and S5G). The relatively low level of neutralizing activity despite high binding titers indicates that the hyperglycosylated monohead nanoparticle elicited a high proportion of non-NAbs, likely targeting the trimer interface. Interestingly, glycan masking of antigens has been shown to alter where on the antigen the majority of responses are being directed but does not change the overall magnitude of the response.26,49 By contrast in this study, the hyperglycosylated monohead mosaic outperformed its non-hyperglycosylated comparator group in every comparison, including overall binding titers across the strains tested, although not always with statistical significance. Alongside the hyperglycosylated monohead, the hyperglycosylated trihead group had the highest mismatched HAI responses against most strains tested (Iowa43, USSR77, and Brisbane07; see Table S2) with the exception of Malaysia54, where the hypervariable trihead group induced the highest HAI titers. This trend in HAI was mirrored in the microneutralization assays, where the highest neutralizing titers were obtained from the two hyperglycosylated trihead groups at weeks 6 and 22 for Malaysia54 and at week 6 for SI06. For the mismatched viruses FM4–7 and USSR77, neutralizing responses were low across all groups at week 6, with the exception of measurable activity in the hypervariable trihead group. However, by week 22, all trihead groups neutralized these viruses more potently than all monohead groups. Taken together, the consistently high vaccine-mismatched HAI and neutralization obtained from the two hyperglycosylated trihead groups suggests that stable trihead closure and hyperglycosylation both contributed toward eliciting superior immune responses.
Epitope mapping of vaccine-elicited antibody responses
We next sought to determine the epitope specificities of the serum antibodies elicited by each vaccine (Figure 5A). We first compared RBS knockout probes NC99-L194W and NC99-T155N/K157T to wild-type NC99 as ELISA antigens to assess the fraction of the antibody response in each group directed at the RBS. NC99-L194W mutates a key highly conserved residue within the RBS pocket, introducing steric bulk that knocks out only antibodies that bind directly to the sialic acid binding pocket, while NC99-T155N/K157T introduces an N-linked glycan that more broadly prevents binding of RBS-directed antibodies.33 Serum antibody binding to both RBS knockout probes was considerably lower than to wild-type NC99 in all trihead groups (binding ratios of 0.08–0.64 for NC99-L194W and 0.03–0.65 for NC99-T155N/K157T), while the differences in relative binding in the monohead groups was smaller (binding ratios of 0.23–1.07 and 0.19–1.07, respectively) (Figures 5B and 5C). We attribute the greater reduction in binding using the T155N/K157T probe compared to the L194W to the fact that the glycan knockin probe will interfere with antibodies targeting a larger area of the antigen. These results clearly indicate that the trihead immunogens elicited substantially more RBS-directed responses than the monohead immunogens.
Figure 5. Epitope mapping of vaccine-elicited antibody responses.
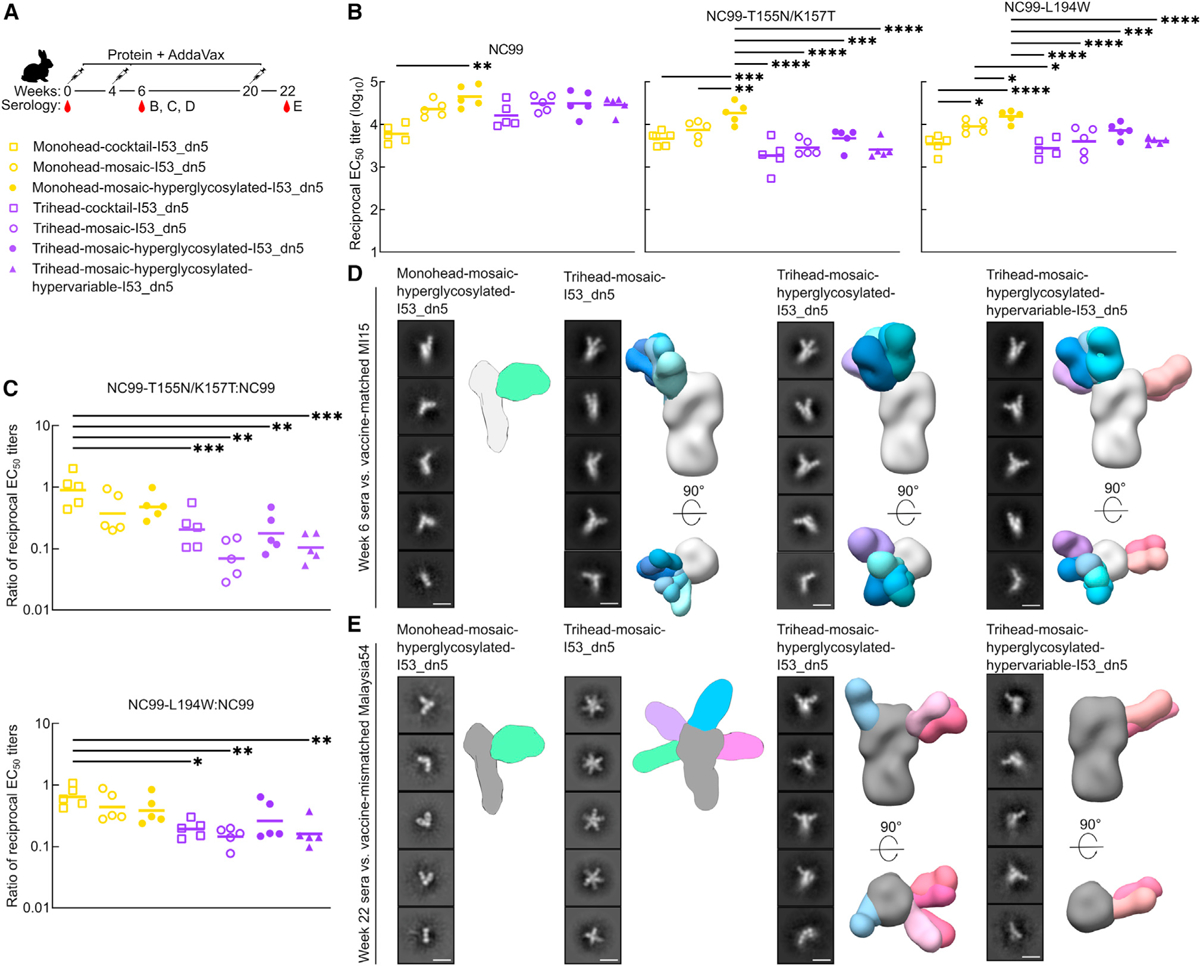
(A) Hypervariable trihead nanoparticle rabbit immunization schedule and groups.
(B) ELISAs using NC99 probes against week 6 rabbit study serum. NC99 ELISA is the same as in Figure 4B. Each symbol represents an individual animal, and the geometric mean of each group is indicated by the bar (n = 5 rabbits/group).
(C) Ratio of NC99 probes to NC99 binding titers in (B).
(D) Representative 2D class averages of week 6 serum from four groups in rabbit study against strain-matched MI15. Hyperglycosylated monohead group has a cartoon schematic of a likely 3D model, while all other groups are composite 3D models of ns-EMPEM analysis.
(E) Representative 2D class averages of week 22 serum from four groups in rabbit study against strain-mismatched Malaysia54. Hyperglycosylated monohead and mosaic trihead groups have a cartoon schematic of their likely 3D models, while other groups are composite 3D models of ns-EMPEM analysis. Scale bars: 15 nm.
Statistical significance was determined using one-way ANOVA with Tukey’s multiple comparisons test; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
We also used nsEM polyclonal epitope mapping (ns-EM-PEM)50,51 to directly visualize where vaccine-elicited serum antibodies bound HA. MI15 strain-matched ns-EMPEM using week 6 serum only detected a small fraction of the antibodies elicited by the hyperglycosylated monohead mosaic bound to the RBS of intact HA trimers, with most Fabs observed to be bound to HA monomers (Figure 5D). A substantial amount of these Fabs likely bind the trimer interface, which has been shown to disrupt trimerization, and indeed our class averages closely resemble those originally reported for FluA-20.21 By contrast, all three mosaic trihead immunogens elicited antibodies that bound mostly to the RBS of intact HA trimers, though a small fraction of Fab bound to monomeric HA was also observed in these groups. We were able to discern RBS-targeting antibody classes with several slightly different angles of approach. Notably, the hypervariable trihead group also exhibited density for Fab binding to the side of the MI15 HA head. This may be attributed to the lack of a glycan in TH-PR34–7gly, TH-MI15–6gly, and TH-SC18–6gly near residues 119–122 (SC18 numbering), in addition to the low occupancy of the N-linked glycan introduced at position 171 in TH-NC99, resulting in inefficient masking at this site. We also performed ns-EMPEM against the vaccine-mismatched Malaysia54 HA using week 22 sera to determine which epitopes were targeted by cross-reactive vaccine-elicited antibodies. We again observed predominantly Fab-bound HA monomers in the hyperglycosylated monohead group, indicating a high proportion of trimer-interface-directed responses (Figure 5E). Week 22 trihead mosaic serum also revealed only Fab-bound HA monomers, but in contrast to the hyperglycosylated monohead group, these Fabs targeted a multitude of various epitopes in the RBD. Finally, in both the hyperglycosylated and hypervariable mosaic trihead sera, we observed Fabs bound only to trimeric Malaysia54 HA, mostly targeting the side of the head, in contrast to the predominantly RBS-directed antibodies seen in these sera at week 6 against the vaccine-matched MI15.
We draw several conclusions from these RBS knockout probe binding and ns-EMPEM data. First, both sets of data suggest that monomeric RBD antigens elicit a high proportion of trimer-interface-directed antibodies, while the closed, stabilized trihead antigens elicit antibodies predominantly targeting the RBS. Second, the week 22 mismatched ns-EMPEM suggests that hyperglycosylation further decreases the elicitation of trimer-interface-directed antibodies by trihead immunogens. Third, the cross-reactive responses obtained with the monohead antigens were largely derived from trimer-interface-directed antibodies, explaining why vaccine-mismatched neutralization is low. Finally, the cross-reactive responses obtained with the hyperglycosylated trihead immunogens predominantly target a site on the side of the RBD near the previously identified lateral patch36 rather than the RBS, especially for the hypervariable immunogen that contains numerous mutations in the RBS periphery.
DISCUSSION
Here, we show that the initial NC99 trihead design strategy described in our accompanying article in this issue of Cell Reports33 can be applied to several divergent H1 HAs and that these trihead mutations improve the potency and breadth of vaccine-elicited antibody responses. The ability of design approaches or specific mutations to generalize across viral families is an important criterion in antigen design that has led to the generation of a number of promising antigen platforms. For example, “stabilized-stem” antigens based on influenza HA were made for both group 1 and 2 HAs, and when displayed on ferritin nanoparticle immunogens, they elicited bNAb responses in animals as well as humans.52–57 We recently reported the stabilization of the closed tetrameric state of several different influenza neuraminidases using an approach that was largely inspired by homology-guided mutations.58 For several class I fusion glycoproteins, strategic use of proline mutations to stabilize the prefusion conformation (or destabilize the postfusion conformation) has proven to be a widely applicable design strategy.59 Proline mutations have been key in the generation of antigens that elicit potent NAb responses against several viruses including HIV48,60 and respiratory syncytial virus (RSV)61 and provided an antigen platform that enabled rapid pandemic response vaccines against SARS-CoV-2.62–64 The generalizability and improved immunogenicity we report here and in the accompanying article in this issue of Cell Reports33 establish triheads as a promising new antigen platform for influenza vaccine design.
The higher HAI and NAb responses we obtained with hyperglycosylated antigens are consistent with previous studies that have established hyperglycosylation as an effective antigen design strategy while also raising mechanistic questions that motivate further studies. Our detailed antigenic characterization clearly revealed that several epitopes were successfully masked by the additional glycans in our hyperglycosylated immunogens, reducing the overall peptidic surface that can be targeted by antibodies. Nevertheless, in our rabbit study, the hyperglycosylated monohead immunogen elicited higher binding titers across various H1 strains than either non-hyperglycosylated monohead immunogen (i.e., cocktail or mosaic). Though we did not assess glycan composition, a possible explanation for this discrepancy is that the potential presence of high-mannose glycans in the hyperglycosylated immunogens may drive better antigen trafficking to lymph nodes and B cell follicles, as demonstrated in recent studies from the Irvine lab.65,66 This effect could result in overall increases in the magnitude or quality of the vaccine-elicited antibody response that are independent of any potential redirection of vaccine-elicited antibody responses to target epitopes due to glycan masking. It is likewise possible that the hyperglycosylated immunogens are less susceptible to proteolytic degradation in vivo, which could also increase immunogenicity overall.67,68 These potential mechanisms are also consistent with the antibody responses elicited by our hyperglycosylated and non-hyperglycosylated trihead immunogens where the former elicited higher HAI and neutralizing activity despite effectively no changes in binding antibody titers. It therefore appears that hyperglycosylation “focused” the antibody response on target epitopes near the RBS. Several previous studies have shown that hyperglycosylation tends to increase the proportion—but not the overall magnitude—of on-target antibodies.26,49 In fact, overall vaccine-elicited antibody titers can be reduced by epitope masking.69 One potential explanation for our observation of clear increases in HAI and neutralizing activity may be an increase in overall immunogenicity accompanied by a reduction in off-target antibody responses. Glycan-dependent effects on trafficking and stability may account for the former, while the epitope masking provided by the glycans could account for the latter. Our EMPEM data in aggregate support suppression of non-RBS responses by hyperglycosylation, with the caveat related to under-occupancy at position 171 of the hyperglycosylated NC99 trihead as noted above. In summary, we observed a focusing-like effect from our hyperglycosylated trihead immunogens, but which of several potential mechanisms was primarily responsible remains to be determined. Future studies on the contributions of various mechanisms to the overall response will benefit from the use of systematic series of immunogens like those described here, in the accompanying article in this issue of Cell Reports,33 and in previous work.65,70,71
In contrast to trihead closure and hyperglycosylation, the effects of mosaic nanoparticle display and hypervariable antigen design were less clear in our experiments. In the first study of mosaic nanoparticle immunogens, a significant advantage in eliciting cross-reactive B cell responses was observed when presenting HA RBDs on ferritin nanoparticles in a mosaic array compared to either a cocktail or sequential immunization regimen.30 In this work, we observed more strain specificity in vaccine-matched responses elicited by the monohead cocktail group compared to the monohead mosaic group. However, we did not observe any significant differences between other cocktail vs. mosaic comparisons for either monohead and trihead immunogens. The addition of the hypervariable RBS periphery also did not show any significant differences compared to the hyperglycosylated trihead group. There are several differences between this study and that of Kanekiyo and colleagues that could account for why mosaics were only superior in the latter, including the animal model, HA strain compositions, nanoparticle size and valency, T cell epitope content, monomeric RBDs vs. trihead antigens, and flexible vs. rigid attachment to the nanoparticle scaffold. One noticeable similarity between the two studies is that the cross-neutralizing responses we observed seem to derive from antibodies directed against the side of the HA head that bind an epitope similar to that of 441D6.30 Although it is likely that glycan under-occupancy at this site is partially responsible, it is intriguing to speculate that BCR cross-linking by mosaic nanoparticles could explain the boost in relatively rare cross-reactive antibodies against this epitope in both studies. Additional studies that more rigorously characterize this epitope on the side of the head and the antibody responses elicited against it could inform future vaccine design efforts.
In conclusion, we have shown that trihead nanoparticle immunogens are a promising platform for next-generation influenza vaccine design. Furthermore, our evaluation of multiple layers of immune-focusing techniques provides a roadmap for efforts using this and other antigen platforms to develop safe and effective vaccines against a variety of pathogens.
Limitations of the study
It is important to note that the animals in this study did not have pre-existing influenza immunity. Humans are commonly exposed to influenza by infection and vaccination, which can profoundly influence immune responses to subsequent exposures.22,72,73 The diversity and complexity of human immune histories further complicates the suitability of animal models for vaccine research. In addition, although our analyses begin to characterize the epitopes targeted by cross-reactive antibodies, serum-based analyses do not provide a complete picture. Studies of vaccine-elicited B cells and mAbs will be required to understand which immunogens most efficiently activate cross-reactive B cells and define which epitopes on vaccine-mismatched antigens are targeted by neutralizing antibodies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Neil King (neilking@uw.edu).
Materials availability
All unique and stable materials generated in this study are available from the lead contact under a Materials Transfer Agreement.
Data and code availability
Structural data generated from this study were deposited in public data repositories and the accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| C05 | Produced in house (Ekiert et al.)6 | N/A |
| 5J8 | Produced in house (Krause et al.)9 | N/A |
| anti-PR34 | Sino Biological | Cat# 11684-R016; RRID:AB_2860374 |
| FluA-20 | Produced in house (Bangaru et al.)21 | N/A |
| Ab6649 | Produced in house (Raymond et al.)36 | N/A |
| Horse anti-mouse IgG, HRP | Cell Signaling Technology | Cat# 7076S; RRID:AB_330924 |
| Goat anti-rabbit IgG, HRP | Cell Signaling Technology | Cat# 7074S; RRID:AB_2099233 |
| Goat anti-human IgG-HRP | Southern Biotech | Cat# 2040–05; RRID:AB_2795644 |
| Bacterial and virus strains | ||
| Influenza reporter viruses | Produced in house (Creanga et al.)80 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| AddaVax adjuvant | InvivoGen | Cat# vac-adx-10 |
| TMB | SeraCare | Cat# 5120-0083 |
| Papain | Thermo Fisher | Cat# 20341 |
| rProtein A Sepharose fast flow resin | Cytiva | Cat# 17127901 |
| Polyethylenimine (PEI) HCl MAX, Linear, Mw 40,000 | Polysciences | Cat# 24765-1 |
| PNGase F | NEB | Cat# P0705 |
| RDE II | VWR | Cat# 10753-482 |
| Deposited data | ||
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40787 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40788 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40792 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40793 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40794 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40795 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40798 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40800 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40801 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40802 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40831 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40832 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40833 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40834 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40835 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40836 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40837 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40838 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40839 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40840 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40841 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40842 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40843 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_ dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40844 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_ dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40845 |
| Experimental models: Cell lines | ||
| Expi293F | ThermoFisher | Cat# A14527 |
| MDCK-SIAT-PB1 | Produced in house (Creanga et al.)80 | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c mice | Jackson Laboratory | Cat# 000651 |
| NZW rabbits | Labcorp, Inc. | https://www.labcorp.com/ |
| Recombinant DNA | ||
| See Table S1 for amino acid sequences | ||
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| EPU | Thermo Fisher | https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/epu-software.html |
| CryoSparc | (Punjani et al.)76 | https://cryosparc.com/ |
| UCSF Chimera | (Pettersen et al.)81 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX | (Goddard et al.)82 | https://www.rbvi.ucsf.edu/chimerax/ |
| PyMol | Schrodinger, LLC | https://pymol.org/2/ |
| RosettaScripts “sugarcoat” code | (Adolf-Bryfogle et al.)34 | N/A |
| Unicorn 7.0 | GE Healthcare | http://www.gelifesciences.com/ |
| Other | ||
| 300 mesh grids | Ted Pella | Cat# 01843-F |
| Filter paper | Cytiva | Cat# 1004047 |
| Uranyl formate | SPI Chem | Cat# 02545-AA |
| Superdex 200 Increase SEC column | Cytiva | Cat# 28-9909-44 |
| Superose 6 Increase SEC column | Cytiva | Cat# 29091596 |
| Talon resin | TaKaRa | Cat# 635652 |
| Excel resin | Cytiva | Cat# 17371203 |
| Isoflurane | USP Patterson | Cat# 07-893-1389 |
| EndoSafe LAL Test Cartridges | Charles River Labs | Cat# PTS20005F |
| Lemo21(DE3) | New England BioLabs | Cat# C2528J |
| MAX Efficiency™ DH5α Competent Cells | Sigma-Aldrich | Cat# I6758 |
| Isopropyl-B-D-thiogalactoside (IPTG) | Thermo Fisher | Cat# 18258012 |
| Kanamycin Sulfate | Sigma-Aldrich | Cat# K1876 |
| HisTrap HP | Cytiva | Cat#17-5247-01 |
| Octet Protein A Biosensors | Sartorius | Cat# 18-5010 |
| Turkey Red Blood Cells | Lampire | Cat# 7249409 |
| Plasmid Plus Maxi Kit | Qiagen | Cat# 12963 |
| Trypsin, TPCK Treated | Thermo Fisher | Cat# 20233 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Expi293F cells are derived from the HEK293F cell line (Life Technologies). Expi293F cells were grown in Expi293 Expression Medium (Life Technologies), cultured at 36.5°C with 8% CO2 and shaking at 150 rpm.
Mice
Female BALB/c mice (Stock # 000,651, BALB/c cByJ mice) four weeks old were obtained from Jackson Laboratory, Bar Harbor, Maine, and maintained at the Comparative Medicine Facility at the University of Washington, Seattle, WA, accredited by the American Association for the Accreditation of Laboratory Animal Care International (AAALAC). Animal procedures were performed under the approvals of the Institutional Animal Care and Use Committee (IACUC) of the University of Washington, Seattle, WA. Only female mice were used to enable evaluation of the immunogens in a consistent model immune system. As a result, we did not evaluate sex as a biological variable.
Rabbits
Female New Zealand white rabbits weighing approximately 4 kg were sourced and housed at Labcorp, Inc. (Denver, PA, USA) and immunizations were performed under permits with approval number COVQ34665P. All immunization procedures complied with all relevant ethical regulations and protocols of the Labcorp Institutional Animal Care and Use Committee.
METHOD DETAILS
Gene expression and Protein purification
All HA constructs used in this study were codon-optimized for human cell expression and made in the CMV/R vector74 by Genscript with a C-terminal hexahistidine affinity tag. PEI MAX was used for transient transfection of HEK293F cells. After four days, mammalian cell supernatants were clarified via centrifugation and filtration. Monohead and trihead components and HA foldons were all purified using IMAC. 1 mL of Ni2+-sepharose Excel or Talon resin was added per 100 mL clarified supernatant along with 5 mL of 1 M Tris, pH 8.0 and 7 mL of 5 M NaCl and left to batch bind while shaking at room temperature for 30 min. Resin was then collected in a gravity column, washed with 5 column volumes of 50 mM Tris, pH 8.0, 500 mM NaCl, 20 mM imidazole, and protein was eluted using 50 mM Tris, pH 8.0, 500 mM NaCl, 300 mM imidazole. Further component purification was done using SEC on a Superdex 200 Increase 10/300 gel filtration column equilibrated in 25 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol. HA-ferritin nanoparticles used in HAI assays were purified using lectin affinity chromatography, followed by SEC on a Superose 6 Increase 10/300 gel filtration column.75
Expression and purification of the I53_dn5A pentamer component from E. coli was carried out by IPTG induction and IMAC followed by SEC on a Superdex 200 Increase 10/300 gel filtration column, respectively.31 Assembly of trihead-I53_dn5 nanoparticles was carried out by mixing purified HA trihead-I53_dn5B and pentameric I53_dn5A components together in vitro at a 1:1 M ratio at 15–40 μM final concentrations. Nanoparticles were left to assemble for 30 min at room temperature with rocking. Nanoparticles were then purified using SEC on a Superose 6 Increase 10/300 gel filtration column equilibrated in 25 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol.
Following purification, nanoparticle quality and concentration was first measured by UV-vis spectroscopy. Nanoparticle polydispersity and purity was then assessed using SDS-PAGE, DLS, and nsEM. Finally, endotoxin levels were measured using the LAL assay, with all immunogens used in animal studies containing less than 100 EU/mg in the final dose. Final immunogens were flash-frozen using liquid nitrogen and stored at −80°C.
Bio-layer interferometry (BLI)
BLI was carried out using an Octet Red 96 system, at 25°C with 1000 rpm shaking. Anti-HA antibodies were diluted in kinetics buffer (PBS with 0.5% serum bovine albumin and 0.01% Tween) to a final concentration of 10 μg/mL before loading onto protein A biosensors (Sartorius) for 200 s. Monohead and trihead components were diluted to 500 nM in kinetics buffer and their association was measured for 200 s, followed by dissociation for 200 s in kinetics buffer alone.
Sandwich BLI
AR2G biosensors (Sartorius) were first activated by the addition of freshly mixed 20 mM 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide and 10 mM Sulfo-N-hydroxysulfosuccinimide. C05 mAb at 5 μg/mL in 10 mM acetate buffer, pH 5.0 was then loaded, followed by quenching with 1 M ethanolamine, pH 8.5. Nanoparticles at 30 μg/mL in kinetics buffer (PBS with 0.5% serum bovine albumin and 0.01% Tween) were then loaded, followed by a baseline step in kinetics buffer before subsequent association and dissociation of anti-PR34 and then 5J8, both at 50 nM in kinetics buffer.
Negative stain electron microscopy 3.5 μL of 70 μg/mL nanoparticles were applied to glow-discharged 400-mesh carbon-coated grids (Electron Microscopy Sciences) and stained with 2% (w/v) uranyl formate. Data were collected using EPU 2.0 on a 120 kV Talos L120C transmission electron microscope (Thermo Scientific) with a BM-Ceta camera. CryoSPARC76 was used for CTF correction, particle picking and extraction, and 2D classification.
Dynamic light scattering
DLS was carried out on an UNcle (UNchained Labs) at 25°C. 10 acquisitions of 5 s each were acquired for each spectrum. Protein concentration (ranging from 0.1–1 mg/mL) and buffer conditions were accounted for in the software.
Glycan occupancy quantitation
Glycan occupancy quantitation was performed using peptide mass spectrometry to determine the relative abundance of peptides in the non-glycosylated and de-glycosylated states after full deglycosylation using PNGaseF.38 Each construct was combined with guanidine hydrochloride and DTT (6 M and 20 mM final concentration, respectively) and boiled for 30 min. Cysteines were then alkylated with the addition of 40 mM iodoacetamide and incubated in the dark for 1 h and quenched with another addition of 20 mM DTT. Samples were diluted 6-fold in 10 mM Tris pH 8.0, 1 mM CaCl2 and treated with PNGaseF (New England Biolabs) for 1 h at 37°C.
The samples were then split into two and treated with either LysC or GluC proteases (ThermoScientific, 1:20 protease:substrate molar ratio) overnight at 37°C. Digestions were quenched with the addition of 0.25% formic acid. Peptides were trapped and desalted using C18 spin columns (ThermoScientific) using the manufacturer’s suggested protocol and dried by speedvac. Purified peptides were resuspended in 20 μL of 0.1% formic acid. LC-MS analysis was performed on an Thermo EASYnLC coupled to a Thermo Orbi-trap Fusion operating in data-dependent mode using EThcD fragmentation. Peptides were resolved over a pulled 30 μm ID silica tip packed with Reprosil-Pur 120 C18-AQ, 5 μm (ESI Source Solutions) using a linear gradient of 5–30%B (A: 0.1% formic acid; B: acetonitrile with 0.1% formic acid) over 90 min at a flowrate of 300 nL/min. LC-MS data were analyzed with Byonic (Protein Metrics Inc.) with a score cutoff of 100 and quantitative analysis was performed using Skyline.77
Circular dichroism
CD measurements were carried out on a JASCO J1500 spectrometer at 25°C, using 1 mm path-length cuvette, at wavelengths from 200 to 260 nm. Proteins were measured at 0.2–0.3 mg/mL in TBS buffer.
Hydrogen deuterium exchange mass spectrometry
40 pmol of each protein per timepoint were incubated in deuterated buffer (20 mM PBS, 85% D2O, pH* 7.48) for 3 s, 1 min, 30 min, and 22 h at room temperature (23°C). The reaction was stopped by diluting 1:1 in ice-cold quench buffer (200 mM tris(2-chlorethyl) phosphate (TCEP), 4 M urea, 0.2% formic acid) to a final pH of 2.495. Samples were immediately flash frozen in liquid nitrogen and stored at 80°C prior to analysis. Online nepenthesin-2 (Nepenthesin-2 protease column, 2.1 × 20 mm, purchased from AffiPro) digestion was performed and analyzed by LC-MS-IMS utilizing a Waters ACQUITY UPLC CSH C18 VanGuard column (130Å, 1.7 μm, 1 mm × 100 mm) and a Waters Synapt G2-Si Q-TOF mass spectrometer.78 A custom HDX cold box maintained the protease digestion at 4°C and the LC plumbing at 0°C throughout the 15 min gradient.79 Waters MSe data were collected on the Waters Synapt G2-Si Q-TOF and were processed using Byonic (Version 3.8, Protein Metrics Inc.) to obtain a peptide reference list for each construct. Some homologous peptides were determined by aligning sequences using Clustal-Omega and calculating theoretical monoisotopic m/z values using ExPASy PeptideMass. Percent exchange values were calculated with theoretical total deuteration profiles produced by HD-Examiner (version 3.3, Sierra Analytics). An internal exchange standard (Pro-Pro-Pro-Ile [PPPI]) was included in each reaction to control for variations in ambient temperature during the labeling reactions. Data for each timepoint was collected in duplicate and error bars were plotted using one standard deviation. Back-exchange was determined by comparing experimental totally deuterated spectra to theoretical totally deuterated spectra. The average back exchange across all peptides was determined to be 31.6%. TH-NC99–9gly data were collected separately from the rest of the triheads and at a higher cone voltage (150 V) which reduced peptide signal at higher charge states (3+, 4+). To control for increases in back exchange from the higher cone voltage, a 3s and 1m control (TH-SC18-RBS2) was run at both voltages (40V and 150V) producing an average difference of 2.6% which is smaller than the differences we observe in the data.
NanoDSF
All proteins were formulated at 0.5 mg/mL in 25 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol and then mixed at 9 volumes to 1 volume of 200× concentrate SYPRO orange (Thermo Fisher) diluted in the same buffer. NanoDSF to determine melting temperatures was carried out on an UNcle (UNchained Labs) by measuring the integration of fluorescence emission spectrum during a thermal ramp from 25°C to 95°C, with a 1°C increase in temperature per minute.
HA sequence conservation Plot
643 unique H1 HA sequences were downloaded from the Influenza Research Database (https://legacy.fludb.org/). Sequence conservation amongst these was then plotted on HA using PyMOL.
(The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LCC).
Immunization
Mice were immunized intramuscularly with 1.5 μg purified nanoparticle immunogen in 100 μL (50 μL in each hind leg) of 50% (v/v) mixture of AddaVax adjuvant (Invivogen, San Diego, CA) (Figures 1 and S1). For sera collection, mice were bled via submental venous puncture 2 weeks following each immunization. Serum was isolated from hematocrit via centrifugation at 2,000 g for 10 min, and stored at −80°C until use. Rabbits were immunized intramuscularly with 25 μg purified nanoparticle immunogen in 1 mL (500 μL in each quadricep) of 50% (v/v) mixture of AddaVax adjuvant (Figures 4, 5, and S5). Rabbits were bled on the day of the first immunization and at 2 weeks following each immunization.
ELISA
HA-foldon trimers were added to 96-well Nunc MaxiSorp plates (Thermo Scientific) at 5.0 μg/mL with 50 μL per well and incubated for 1 h. Blocking buffer composed of Tris-Buffered Saline Tween (TBST: 25 mM Tris pH 8.0, 150 mM NaCl, 0.05% (v/v) Tween 20) with 5% Nonfat milk was then added at 200 μL per well and incubated for 1 h. Next plates were washed, with all washing steps consisting of 3× washing with TBST using a robotic plate washer (Biotek). 5-fold serial dilutions of serum starting at 1:100 were made in blocking buffer, added to plates at 50 μL per well, and incubated for 1 h. Plates were washed again before addition of 50 μL per well of either anti-mouse or anti-rabbit HRP-conjugated goat secondary antibody (CellSignaling Technology) diluted 1:2,000 in blocking buffer and incubated for 30 min. All incubations were carried out with shaking at room temperature. Plates were washed a final time, and then 100 μL per well of TMB (3,3′,5′,5-tetramethylbenzidine, SeraCare) was added for 2 min, followed by quenching with 100 μL per well of 1 N HCl. Reading at 450 nm absorbance was done on an Epoch plate reader (BioTek).
Competition ELISA
Competition ELISAs were performed in the same manner as the above ELISA protocol with some modifications as follows. For competition with FluA-20, 5-fold serial serum dilutions were made starting at 1:10, and for competition with C05, 3-fold serial serum dilutions were made starting at 1:5. Serum was left to incubate for 30 min, followed by addition of 50 μL of 0.1 μg/mL competitor antibody in blocking buffer and incubation for 45 min. After washing, anti-human HRP-conjugated goat secondary antibody (Southern Biotech) was added at 20,000× at 50 μL per well and incubated for 30 min.
HAI
Serum was inactivated using receptor-destroying enzyme (RDE) II (Seiken) in PBS at a 3:1 ratio of RDE to serum for 16 h at 37°C, followed by 40 min at 56°C. Inactivated serum was serially diluted 2-fold in PBS in V-bottom plates at 25 μL per well. 25 μL HA-ferritin nanoparticles at 4 hemagglutinating units were then added to all wells and incubated at room temperature for 30 min.45 Lastly, 50 μL of 10-fold diluted turkey red blood cells (Lampire) in PBS was added to each well. Hemagglutination was left to proceed for at least 1 h before recording HAI titer.
Reporter-based microneutralization assay
Influenza A reporter viruses were made from H1N1 viruses with a modified PB1 segment expressing the TdKatushka reporter gene (R3ΔPB1), rescued, and propagated in MDCK-SIAT-PB1 cells in the presence of TPCK-treated trypsin (1 μg mL−1, Sigma) at 37°C.80 Virus stocks were stored at −80°C and were titrated before use in the assay. Rabbit sera was treated with receptor destroying enzyme (RDE II; Denka Seiken) and heat-inactivated before use in neutralization assays. 384 well plates (Greiner) were pre-seeded with 1.0 × 105 MDCK-SIAT1-PB1 cells and incubated overnight. Immune sera or monoclonal antibody controls (CR8071 and CR911410) were serially diluted and incubated for 1 h at 37°C with pre-titrated virus (A/Fort Monmouth/1/1947, A/Malaysia/302/1954, A/New Caledonia/20/1999, A/USSR/90/1977, A/Solomon Islands/3/2006, A/Michigan/45/2015). Serum-virus mixtures were then transferred in quadruplicate onto the pre-seeded 384 well plates and incubated at 37°C for 18–26 h. The number of fluorescent cells in each well was counted automatically using a Celigo image cytometer (Nexcelom Biosciences). IC50 values, defined as the serum dilution or antibody concentration that gives 50% reduction in virus-infected cells, were calculated from neutralization curves using a four-parameter nonlinear regression model.
EMPEM
1 mL rabbit serum was diluted 3× in PBS and incubated overnight with 1 mL packed rProtein A Sepharose Fast Flow resin (Cytiva). The FT was removed using gravity purification and resin was washed with 20 CVs PBS. IgGs were eluted by incubating resin for 20 min with 1 CV of 0.1 M glycine, pH 2.5, repeated twice. Elutions were neutralized with 1 M Tris, pH 8.0 to a final concentration of 50 mM. IgGs were then buffer exchanged into PBS and concentrated to 250 μL for digestion into Fabs. Fab digestion was carried out by adding in 250 μL freshly made 2× digestion buffer (40 mM NaPO4 pH 6.5, 20 mM EDTA, 40 mM Cysteine) and 500 μL papain resin in 500 μL 1× digestion buffer, and incubated with shaking at 37°C for 16 h. Papain digestion reaction was centrifuged and supernatant containing fabs was collected and filtered. Papain was subsequently washed with 1 CV 20 mM Tris, pH 8.0 and this supernatant was added to the first. Digested sera was then purified by SEC on a Superdex 200 Increase 10/300 GL column. Purified Fabs were then concentrated to 50 μL, mixed with 50-fold molar excess of HA-foldon trimers, and incubated for 16–20 h at room temperature with gentle rocking. Immune complexes were purified by SEC on a Superdex 200 Increase 10/300 GL column and used in EM. nsEM data were collected using EPU 2.0 on a 120 kV Talos L120C transmission electron microscope (Thermo Scientific) with a BM-Ceta camera. Data processing was done in CryoSPARC,76 starting with CTF correction, particle picking and extraction. Three rounds of 2D classification were done, keeping only classes that had either HA monomer or trimer with bound Fabs. 3D models for these immune complexes were then generated using ab initio 3D reconstruction and heterogeneous refinement without imposing any symmetry. Classes that had clear, trimeric HA density and were representative of the diversity of Fab binding in each sample of polyclonal serum without redundancy were then separately subjected to a 3D refinement. Figure models were generated using Chimera81 and ChimeraX.82
Statistical analysis
Multi-group comparisons were performed using the Brown-Forsythe one-way ANOVA test and Dunnett’s T3 post hoc analysis in Prism 9 (GraphPad). Differences were considered significant when p values were less than 0.05.
Supplementary Material
Highlights.
Design of triheads for several H1 strains and hyperglycosylated and hypervariable variants
Trihead trimer-stabilizing mutations reduce non-neutralizing antibodies in mice and rabbits
Hyperglycosylated triheads elicit higher responses against broadly neutralizing epitopes
ACKNOWLEDGMENTS
This work was funded by a generous gift from Open Philanthropy (N.P.K.); the Audacious Project at the Institute for Protein Design (N.P.K.); the Defense Threat Reduction Agency (HDTRA1-18-1-0001 to N.P.K.); the National Institute of Allergy and Infectious Diseases (P01 AI167966 to N.P.K.); and the intramural research program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (M.K.). This work was supported in part by the University of Washington’s Proteomics Resource (UWPR95794) and the UW Molecular Biophysics Training Program (T32GM008268). Labcorp performed all rabbit immunizations and blood draws.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113553.
DECLARATION OF INTERESTS
N.P.K. is a co-founder, shareholder, paid consultant, and chair of the scientific advisory board of Icosavax, Inc. The King lab has received unrelated sponsored research agreements from Pfizer and GSK. D.E. is a shareholder of Icosavax, Inc. A.D., D.E., M.K., and N.P.K. are listed as co-inventors on patent applications filed by the University of Washington related to this work.
REFERENCES
- 1.Carrat F, and Flahault A (2007). Influenza vaccine: the challenge of antigenic drift. Vaccine 25, 6852–6862. [DOI] [PubMed] [Google Scholar]
- 2.Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, and Rambaut A (2014). Integrating influenza antigenic dynamics with molecular evolution. Elife 3, e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GCM, Vervaet G, Skepner E, Lewis NS, Spronken MIJ, Russell CA, et al. (2013). Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979. [DOI] [PubMed] [Google Scholar]
- 4.Rejmanek D, Hosseini PR, Mazet JAK, Daszak P, and Goldstein T (2015). Evolutionary Dynamics and Global Diversity of Influenza A Virus. J. Virol 89, 10993–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu NC, and Wilson IA (2017). A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus. J. Mol. Biol 429, 2694–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, et al. (2012). Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 108, 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, and Wilson IA (2012). Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. USA 109, 17040–17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, and Crowe JE Jr. (2011). A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol 85, 10905–10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. (2016). Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166, 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Chen J, Zheng Q, Xue W, Zhang L, Rong R, Zhang S, Wang Q, Hong M, Zhang Y, et al. (2022). Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses. Nat. Commun 13, 5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, and Harrison SC (2018). Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 48, 174–184.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altman MO, Bennink JR, Yewdell JW, and Herrin BR (2015). Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. Elife 4, e07467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. (2017). Defining B cell immunodominance to viruses. Nat. Immunol 18, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altman MO, Angeletti D, and Yewdell JW (2018). Antibody Immunodominance: The Key to Understanding Influenza Virus Antigenic Drift. Viral Immunol. 31, 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zost SJ, Lee J, Gumina ME, Parkhouse K, Henry C, Wu NC, Lee C-CD, Wilson IA, Wilson PC, Bloom JD, and Hensley SE (2019). Identification of Antibodies Targeting the H3N2 Hemagglutinin Receptor Binding Site following Vaccination of Humans. Cell Rep. 29, 4460–4470.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthmiller JJ, Han J, Utset HA, Li L, Lan LY-L, Henry C, Stamper CT, McMahon M, O’Dell G, Fernández-Quintero ML, et al. (2022). Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Nature 602, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benton DJ, Nans A, Calder LJ, Turner J, Neu U, Lin YP, Ketelaars E, Kallewaard NL, Corti D, Lanzavecchia A, et al. (2018). Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. USA 115, 10112–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, et al. (2019). Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 177, 1124–1135.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, et al. (2019). A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 177, 1136–1152.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee C-H, Lavinder JJ, et al. (2016). Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med 22, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y, and Suzuki Y (2012). Evidence for N-glycan shielding of antigenic sites during evolution of human influenza A virus hemagglutinin. J. Virol 86, 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei C-J, Boyington JC, Dai K, Houser KV, Pearce MB, Kong W-P, Yang Z-Y, Tumpey TM, and Nabel GJ (2010). Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med 2, 24ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eggink D, Goff PH, and Palese P (2014). Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol 88, 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, and Schmidt AG (2019). Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 25, 827–835.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thornlow DN, Macintyre AN, Oguin TH, Karlsson AB, Stover EL, Lynch HE, Sempowski GD, and Schmidt AG (2021). Altering the Immunogenicity of Hemagglutinin Immunogens by Hyperglycosylation and Disulfide Stabilization. Front. Immunol 12, 737973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun W, Kirkpatrick E, Ermler M, Nachbagauer R, Broecker F, Krammer F, and Palese P (2019). Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol 93, e003333–e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broecker F, Liu STH, Suntronwong N, Sun W, Bailey MJ, Nachbagauer R, Krammer F, and Palese P (2019). A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. (2019). Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol 20, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyoglu-Barnum S, Ellis D, Gillespie RA, Hutchinson GB, Park Y-J, Moin SM, Acton OJ, Ravichandran R, Murphy M, Pettie D, et al. (2021). Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen AA, Yang Z, Gnanapragasam PNP, Ou S, Dam K-MA, Wang H, and Bjorkman PJ (2021). Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS One 16, e0247963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis D, Dosey A, Boyoglu-Barnum S, Park Y-J, Gillespie R, Syeda H, Tsybovsky Y, Murphy M, Carter L, Ueda G, et al. Antigen-antigen Spacing on Protein Nanoparticles Influences Antibody Responses to Vaccination. [DOI] [PMC free article] [PubMed]
- 34.Adolf-Bryfogle J, Labonte JW, Kraft JC, Shapovalov M, Raemisch S, Lütteke T, DiMaio F, Bahl CD, Pallesen J, King NP, et al. (2021). Growing Glycans in Rosetta: Accurate de novo glycan modeling, density fitting, and rational sequon design. Preprint at bioRxiv, 2021.09.27. 462000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, Alam N, Alford RF, Aprahamian M, Baker D, Barlow KA, Barth P, et al. (2020). Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods 17, 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, and Harrison SC (2018). Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 115, 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Struwe WB, Stuckmann A, Behrens A-J, Pagel K, and Crispin M (2017). Global N-Glycan Site Occupancy of HIV-1 gp120 by Metabolic Engineering and High-Resolution Intact Mass Spectrometry. ACS Chem. Biol 12, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Varón Silva D, Fuchser J, Kaspar S, Rapp E, Seeberger PH, and Kolarich D (2013). Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom 48, 627–639. [DOI] [PubMed] [Google Scholar]
- 39.Cohen AA, Gnanapragasam PNP, Lee YE, Hoffman PR, Ou S, Kakutani LM, Keeffe JR, Wu H-J, Howarth M, West AP, et al. (2021). Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walls AC, Miranda MC, Schäfer A, Pham MN, Greaney A, Arunachalam PS, Navarro M-J, Tortorici MA, Rogers K, O’Connor MA, et al. (2021). Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell 184, 5432–5447.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sliepen K, Radić L, Capella-Pujol J, Watanabe Y, Zon I, Chumbe A, Lee W-H, de Gast M, Koopsen J, Koekkoek S, et al. (2022). Induction of cross-neutralizing antibodies by a permuted hepatitis C virus glycoprotein nanoparticle vaccine candidate. Nat. Commun 13, 7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen AA, van Doremalen N, Greaney AJ, Andersen H, Sharma A, Starr TN, Keeffe JR, Fan C, Schulz JE, Gnanapragasam PNP, et al. (2022). Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 377, eabq0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brinkkemper M, Veth TS, Brouwer PJM, Turner H, Poniman M, Burger JA, Bouhuijs JH, Olijhoek W, Bontjer I, Snitselaar JL, et al. (2022). Co-display of diverse spike proteins on nanoparticles broadens sarbecovirus neutralizing antibody responses. iScience 25, 105649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martín J, Wharton SA, Lin YP, Takemoto DK, Skehel JJ, Wiley DC, and Steinhauer DA (1998). Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology 241, 101–111. [DOI] [PubMed] [Google Scholar]
- 45.Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. (2014). Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol 88, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doud MB, and Bloom JD (2016). Accurate Measurement of the Effects of All Amino-Acid Mutations on Influenza Hemagglutinin. Viruses 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu NC, Xie J, Zheng T, Nycholat CM, Grande G, Paulson JC, Lerner RA, and Wilson IA (2017). Diversity of Functionally Permissive Sequences in the Receptor-Binding Site of Influenza Hemagglutinin. Cell Host Microbe 21, 742–753.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. (2015). HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duan H, Chen X, Boyington JC, Cheng C, Zhang Y, Jafari AJ, Stephens T, Tsybovsky Y, Kalyuzhniy O, Zhao P, et al. (2018). Glycan Masking Focuses Immune Responses to the HIV-1 CD4-Binding Site and Enhances Elicitation of VRC01-Class Precursor Antibodies. Immunity 49, 301–311.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, Bastidas R, Nedellec R, McCoy LE, Wilson IA, et al. (2018). Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 49, 288–300.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han J, Schmitz AJ, Richey ST, Dai Y-N, Turner HL, Mohammed BM, Fremont DH, Ellebedy AH, and Ward AB (2021). Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Cell Rep. 34, 108682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yassine HM, Boyington JC, McTamney PM, Wei C-J, Kanekiyo M, Kong W-P, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. (2015). Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med 21, 1065–1070. [DOI] [PubMed] [Google Scholar]
- 53.Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, et al. (2019). Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moin SM, Boyington JC, Boyoglu-Barnum S, Gillespie RA, Cerutti G, Cheung CS-F, Cagigi A, Gallagher JR, Brand J, Prabhakaran M, et al. (2022). Co-immunization with hemagglutinin stem immunogens elicits cross-group neutralizing antibodies and broad protection against influenza A viruses. Immunity 55, 2405–2418.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews SF, Cominsky LY, Shimberg GD, Gillespie RA, Gorman J, Raab JE, Brand J, Creanga A, Gajjala SR, Narpala S, et al. (2023). An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans. Sci. Transl. Med 15, eade4976. [DOI] [PubMed] [Google Scholar]
- 56.Darricarrère N, Qiu Y, Kanekiyo M, Creanga A, Gillespie RA, Moin SM, Saleh J, Sancho J, Chou T-H, Zhou Y, et al. (2021). Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med 13, eabe5449. [DOI] [PubMed] [Google Scholar]
- 57.Widge AT, Hofstetter AR, Houser KV, Awan SF, Chen GL, Burgos Florez MC, Berkowitz NM, Mendoza F, Hendel CS, Holman LA, et al. (2023). An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci. Transl. Med 15, eade4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellis D, Lederhofer J, Acton OJ, Tsybovsky Y, Kephart S, Yap C, Gillespie RA, Creanga A, Olshefsky A, Stephens T, et al. (2022). Structure-based design of stabilized recombinant influenza neuraminidase tetramers. Nat. Commun 13, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders RW, and Moore JP (2021). Virus vaccines: proteins prefer prolines. Cell Host Microbe 29, 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJM, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, and Langedijk JPM (2015). A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat. Commun 6, 8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, et al. (2017). Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 114, E7348–E7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh C-L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H-C, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, et al. (2020). Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, and McLellan JS (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Read BJ, Won L, Kraft JC, Sappington I, Aung A, Wu S, Bals J, Chen C, Lee KK, Lingwood D, et al. (2022). Mannose-binding lectin and complement mediate follicular localization and enhanced immunogenicity of diverse protein nanoparticle immunogens. Cell Rep. 38, 110217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tokatlian T, Read BJ, Jones CA, Kulp DW, Menis S, Chang JYH, Steichen JM, Kumari S, Allen JD, Dane EL, et al. (2019). Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 363, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. (2019). Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 177, 1153–1171.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aung A, Cui A, Maiorino L, Amini AP, Gregory JR, Bukenya M, Zhang Y, Lee H, Cottrell CA, Morgan DM, et al. (2023). Low protease activity in B cell follicles promotes retention of intact antigens after immunization. Science 379, eabn8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weidenbacher PA, and Kim PS (2019). Protect, modify, deprotect (PMD): A strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc. Natl. Acad. Sci. USA 116, 9947–9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kato Y, Abbott RK, Freeman BL, Haupt S, Groschel B, Silva M, Menis S, Irvine DJ, Schief WR, and Crotty S (2020). Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 53, 548–563.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cobey S, and Hensley SE (2017). Immune history and influenza virus susceptibility. Curr. Opin. Virol 22, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andrews SF, Raab JE, Gorman J, Gillespie RA, Cheung CSF, Rawi R, Cominsky LY, Boyington JC, Creanga A, Shen C-H, et al. (2022). A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination. Nat. Med 28, 373–382. [DOI] [PubMed] [Google Scholar]
- 74.Barouch DH, Yang Z-Y, Kong W-P, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, et al. (2005). A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol 79, 8828–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanekiyo M, Wei C-J, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, Rao SS, Kong W-P, Wang L, and Nabel GJ (2013). Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryo-SPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- 77.Pino LK, Searle BC, Bollinger JG, Nunn B, MacLean B, and Mac-Coss MJ (2020). The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev 39, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verkerke HP, Williams JA, Guttman M, Simonich CA, Liang Y, Filipavicius M, Hu S-L, Overbaugh J, and Lee KK (2016). Epitope-Independent Purification of Native-Like Envelope Trimers from Diverse HIV-1 Isolates. J. Virol 90, 9471–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watson MJ, Harkewicz R, Hodge EA, Vorauer C, Palmer J, Lee KK, and Guttman M (2021). Simple Platform for Automating Decoupled LC-MS Analysis of Hydrogen/Deuterium Exchange Samples. J. Am. Soc. Mass Spectrom 32, 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, Hatch L, Stephens T, Tsybovsky Y, Crank MC, et al. (2021). A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nat. Commun 12, 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 82.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data generated from this study were deposited in public data repositories and the accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| C05 | Produced in house (Ekiert et al.)6 | N/A |
| 5J8 | Produced in house (Krause et al.)9 | N/A |
| anti-PR34 | Sino Biological | Cat# 11684-R016; RRID:AB_2860374 |
| FluA-20 | Produced in house (Bangaru et al.)21 | N/A |
| Ab6649 | Produced in house (Raymond et al.)36 | N/A |
| Horse anti-mouse IgG, HRP | Cell Signaling Technology | Cat# 7076S; RRID:AB_330924 |
| Goat anti-rabbit IgG, HRP | Cell Signaling Technology | Cat# 7074S; RRID:AB_2099233 |
| Goat anti-human IgG-HRP | Southern Biotech | Cat# 2040–05; RRID:AB_2795644 |
| Bacterial and virus strains | ||
| Influenza reporter viruses | Produced in house (Creanga et al.)80 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| AddaVax adjuvant | InvivoGen | Cat# vac-adx-10 |
| TMB | SeraCare | Cat# 5120-0083 |
| Papain | Thermo Fisher | Cat# 20341 |
| rProtein A Sepharose fast flow resin | Cytiva | Cat# 17127901 |
| Polyethylenimine (PEI) HCl MAX, Linear, Mw 40,000 | Polysciences | Cat# 24765-1 |
| PNGase F | NEB | Cat# P0705 |
| RDE II | VWR | Cat# 10753-482 |
| Deposited data | ||
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40787 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40788 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40792 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40793 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40794 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40795 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40798 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40800 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40801 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40802 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40831 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40832 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40833 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40834 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40835 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40836 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40837 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40838 |
| Negative-stain EM map of week 6 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_dn5 in complex with MI15 HA-foldon. | EMDataBank | EMD: 40839 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40840 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40841 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40842 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hyperglycosylated-Mosaic-I53_dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40843 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_ dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40844 |
| Negative-stain EM map of week 22 polyclonal serum from rabbit immunized with Trihead-Hypervariable-Mosaic-I53_ dn5 in complex with Malaysia54 HA-foldon. | EMDataBank | EMD-40845 |
| Experimental models: Cell lines | ||
| Expi293F | ThermoFisher | Cat# A14527 |
| MDCK-SIAT-PB1 | Produced in house (Creanga et al.)80 | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c mice | Jackson Laboratory | Cat# 000651 |
| NZW rabbits | Labcorp, Inc. | https://www.labcorp.com/ |
| Recombinant DNA | ||
| See Table S1 for amino acid sequences | ||
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| EPU | Thermo Fisher | https://www.thermofisher.com/us/en/home/electron-microscopy/products/software-em-3d-vis/epu-software.html |
| CryoSparc | (Punjani et al.)76 | https://cryosparc.com/ |
| UCSF Chimera | (Pettersen et al.)81 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX | (Goddard et al.)82 | https://www.rbvi.ucsf.edu/chimerax/ |
| PyMol | Schrodinger, LLC | https://pymol.org/2/ |
| RosettaScripts “sugarcoat” code | (Adolf-Bryfogle et al.)34 | N/A |
| Unicorn 7.0 | GE Healthcare | http://www.gelifesciences.com/ |
| Other | ||
| 300 mesh grids | Ted Pella | Cat# 01843-F |
| Filter paper | Cytiva | Cat# 1004047 |
| Uranyl formate | SPI Chem | Cat# 02545-AA |
| Superdex 200 Increase SEC column | Cytiva | Cat# 28-9909-44 |
| Superose 6 Increase SEC column | Cytiva | Cat# 29091596 |
| Talon resin | TaKaRa | Cat# 635652 |
| Excel resin | Cytiva | Cat# 17371203 |
| Isoflurane | USP Patterson | Cat# 07-893-1389 |
| EndoSafe LAL Test Cartridges | Charles River Labs | Cat# PTS20005F |
| Lemo21(DE3) | New England BioLabs | Cat# C2528J |
| MAX Efficiency™ DH5α Competent Cells | Sigma-Aldrich | Cat# I6758 |
| Isopropyl-B-D-thiogalactoside (IPTG) | Thermo Fisher | Cat# 18258012 |
| Kanamycin Sulfate | Sigma-Aldrich | Cat# K1876 |
| HisTrap HP | Cytiva | Cat#17-5247-01 |
| Octet Protein A Biosensors | Sartorius | Cat# 18-5010 |
| Turkey Red Blood Cells | Lampire | Cat# 7249409 |
| Plasmid Plus Maxi Kit | Qiagen | Cat# 12963 |
| Trypsin, TPCK Treated | Thermo Fisher | Cat# 20233 |


