SUMMARY
Immunogen design approaches aim to control the specificity and quality of antibody responses elicited by next-generation vaccines. Here, we use computational protein design to generate a nanoparticle vaccine platform based on the receptor-binding domain (RBD) of influenza hemagglutinin (HA) that enables precise control of antigen conformation and spacing. HA RBDs are presented as either monomers or native-like closed trimers that are connected to the underlying nanoparticle by a rigid linker that is modularly extended to precisely control antigen spacing. Nanoparticle immunogens with decreased spacing between trimeric RBDs elicit antibodies with improved hemagglutination inhibition and neutralization potency as well as binding breadth across diverse H1 HAs. Our “trihead” nanoparticle immunogen platform provides insights into anti-HA immunity, establishes antigen spacing as an important parameter in structure-based vaccine design, and embodies several design features that could be used in next-generation vaccines against influenza and other viruses.
Graphical Abstract
In brief
Ellis et al. used computational protein design to create a series of trimeric hemagglutinin-head-based nanoparticles with differing extension domains to vary antigen spacing. They found that decreased antigen spacing leads to a more potent and broad immune response as well as changes in epitope specificity.
INTRODUCTION
Influenza vaccines are of significant biomedical importance, both for the prevention of disease and as tools for studying vaccinology. The viral hemagglutinin (HA) is the key antigen of current influenza vaccines and is a well-studied model antigen. HA is a homotrimeric class I fusion protein responsible for both binding to sialoside receptors on host cells and mediating membrane fusion to deliver viral RNA.1,2 HA consists of two distinct structural domains—the head and the stem—which have distinct functional and immunogenic properties. The head domain, comprising an upper receptor-binding domain (RBD) and a lower vestigial esterase (VE) subdomain, is immunodominant, hypervariable, and the primary target of antibodies elicited by current commercial influenza vaccines.3–6 In contrast, the immunosubdominant stem domain contains the membrane fusion machinery and bears broadly conserved epitopes for antibodies.2 Antibody responses against the HA head often potently neutralize virions by blocking receptor binding, an effect that can be measured using hemagglutination inhibition (HAI), a long-established correlate of protection.7 However, the immunodominance of highly variable head epitopes and biased immune memory induced by early encounters with influenza antigens lead to narrow antibody specificities,8 forcing regular updates to vaccine formulations to keep up with mutations in circulating viruses.8,9 Improving the potency of antibody responses could lead to direct increases in vaccine efficacy.10,11
The HA head has a variety of epitopes that have been extensively studied, revealing distinct protective mechanisms and differences in epitope conservation. Early studies with murine monoclonal antibodies (mAbs) divided the HA head into multiple antigenic sites and showed that sites nearer the sialic acid receptor-binding site (RBS) mutate more frequently.12–15 More recent studies have identified epitopes in the HA head with higher conservation that could be targeted by broadly cross-reactive vaccines. RBS-directed antibodies can broadly neutralize viruses within the same HA subtype16–21—and in some cases sporadically across subtypes22–26—but may require avidity to achieve potency and can be sensitive to escape mutations.17 More recently, interface-directed mAbs recognizing cryptic epitopes in the interface between head domains were shown to be capable of providing broad protection across diverse influenza A subtypes in preclinical studies.27–30 Because these interface-directed mAbs lack neutralizing activity, effector functions are thought to play a key role in their protective capacity. Other subtype-specific epitopes throughout the head have the potential to elicit antibodies with HAI or neutralizing activity, or effector-function-driven immunity.25,29,31–36 The location and angle of approach in part determine the protective mechanisms of head-directed antibodies, with HAI (i.e., receptor blocking) achieved by antibodies targeting apical epitopes or competing directly with binding to sialosides, while neutralizing head-directed antibodies without HAI activity often target epitopes more distant from the RBS. The diversity of epitopes in the HA head and the antibody responses they elicit offer multiple opportunities for engineering the quality of immune responses induced by next-generation vaccines.
Efforts to enhance head-directed antibody responses against specific epitopes have focused on direct modifications to epitopes in the HA head or the way it is presented to the immune system. One approach is to incorporate a diverse collection of epitopes from various viruses in single HA antigens to increase protective breadth through direct inclusion of broadly representative epitopes.37–39 Hyperglycosylation and engineered interprotomer disulfide bonds have been used to redirect antibody responses away from less desirable epitopes and focus them on40 or away from41 the trimer interface. Heterologous prime-boost regimens using chimeric antigens that maintain target epitopes42,43 or, alternatively, diversify off-target epitopes44,45 or entire domains46,47 have also been evaluated as a mechanism for focusing antibody responses on target epitopes. Interestingly, early clinical studies with chimeric HA antigens featuring exotic head domains demonstrated the elicitation of interface-directed antibodies, perhaps because these antigens adopt open conformations that expose this epitope.46,48 Innovative approaches for displaying HA antigens on nanoparticles have also been used to shape polyclonal antibody responses. For example, co-display of multiple HA RBD antigens of H1 subtype on ferritin nanoparticles improved antibody responses against broadly conserved epitopes within H1 HAs.34 Identifying additional strategies to maximize and shape antibody responses against target epitopes on the HA head could contribute to the development of improved influenza vaccines and inspire vaccine design for other pathogens.
The immunogenicity of a given antigen has long been linked to its spatial organization.49 Antigen repetition and the spacing between antigens on virions have been argued to play key roles in antibody responses to infection, explaining why the immune system has evolved to recognize repetition as a danger signal.50–53 Early studies demonstrating the importance of antigen oligomerization in the context of vaccination suggested that repetitive display every 5–15 nm may be ideally immunogenic.54–58 More recent studies have used DNA origami to precisely probe the effects of antigen spacing and valency on antibody avidity and B cell activation in vitro.59,60 Likewise, engineered series of protein nanoparticle immunogens have shown that increasing antigen valency leads to enhanced B cell activation and antibody responses in vivo.61,62 However, defining structural correlates of immunogenicity and applying this knowledge to improve vaccine design have been challenging due to the lack of tools capable of generating systematic series of immunogens that vary specific structural features with high precision.
Here, we describe a protein nanoparticle immunogen platform in which the oligomeric state and the spatial organization of displayed HA RBDs can be precisely controlled. We find that both features contribute to the immunogenicity of various epitopes to shape the overall magnitude and quality of the vaccine-elicited antibody response. Our results suggest new approaches to vaccine development for influenza and other pathogens through precisely tailored nanoparticle immunogen design.
RESULTS
Design and characterization of a trihead nanoparticle immunogen
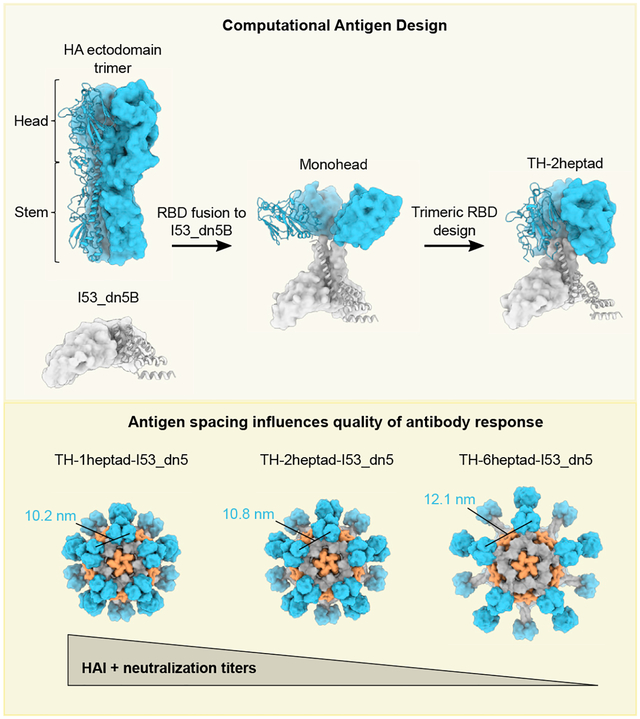
HA naturally forms a trimer with the stem domain containing the majority of interprotomeric contacts, while the RBD region in the head domain has relatively fewer contacts, allowing for some flexibility or “breathing.”63,64 When expressed without the stem domain, HA RBDs are monomeric when fused to other multimeric structures, such as protein nanoparticles.34 We sought to develop structure-based strategies for controlling two aspects of HA RBD antigen display on protein nanoparticles. First, we aimed to conformationally fix HA RBD antigens in either “closed” trimeric or “open” monomeric conformations, respectively hiding or exposing epitopes at the interface between RBDs to determine whether each conformation has distinct immunogenic properties. Second, we aimed to study in detail the impact of antigen organization on immunogenicity by designing nanoparticle immunogens that display HA RBD antigens with precisely defined antigen spacing and rigidity.
As an initial step toward both goals, we aimed to use a trimeric helical bundle as a rigid, extensible linker between the displayed HA RBD antigen and an underlying protein nanoparticle scaffold. Symmetrical helical bundles can be modularly constructed from motifs such as heptad repeats to form rigid units of arbitrary length.65–67 A mutated homotrimeric GCN4-derived helical bundle with C3 symmetry built from such repeats (PDB: 1GCM) was previously observed to closely match the backbone geometry of the apical portion of the HA stem that natively contacts the RBDs.68,69 Furthermore, the C-terminal end of this GCN4 trimer closely matches the N-terminal helices of I53_dn5B, the trimeric component of the computationally designed icosahedral nanoparticle I53_dn5.70 We reasoned that using this helical bundle as a rigid connection between the RBD and I53_dn5B might stabilize the RBDs in a native-like closed trimeric state while also allowing for precise control of antigen spacing by simply modifying the number of heptad repeats.
A preliminary design was constructed by fusing the RBD (residues 56–264) of the H1N1 isolate A/New Caledonia/20/1999 (NC99) to two heptad repeats of the modified trimeric GCN4 bundle and I53_dn5B (Figures 1A and 1B). The N-terminal portion of GCN4 was redesigned to match residues in the HA stem that contact the HA RBD to determine if these interactions and GCN4-mediated trimerization were sufficient to drive trimeric closure of the heads. The C-terminal end of GCN4 was blended into a single continuous helix with the N terminus of I53_dn5B (referred to as the “heptad domain”) by optimally aligning the hydrophobic residues that form the core of the three-helix bundle with those of I53_dn5B. The resultant protein (referred to as “monohead”) was successfully secreted from HEK293F cells, purified by immobilized metal affinity chromatography (IMAC), and confirmed as a trimer by size-exclusion chromatography (SEC) (Figure S1A). Biolayer interferometry (BLI) showed that the monohead component bound well to the RBS-directed mAb C0522 compared to a monomeric NC99-RBD benchmark antigen, suggesting native-like HA RBD folding (Figure 1C). However, it also bound the non-neutralizing trimer-interface-directed antibody FluA-20,29 which demonstrated that the interface region remained exposed and accessible for antibody recognition.
Figure 1. Design and characterization of a trihead nanoparticle immunogen.
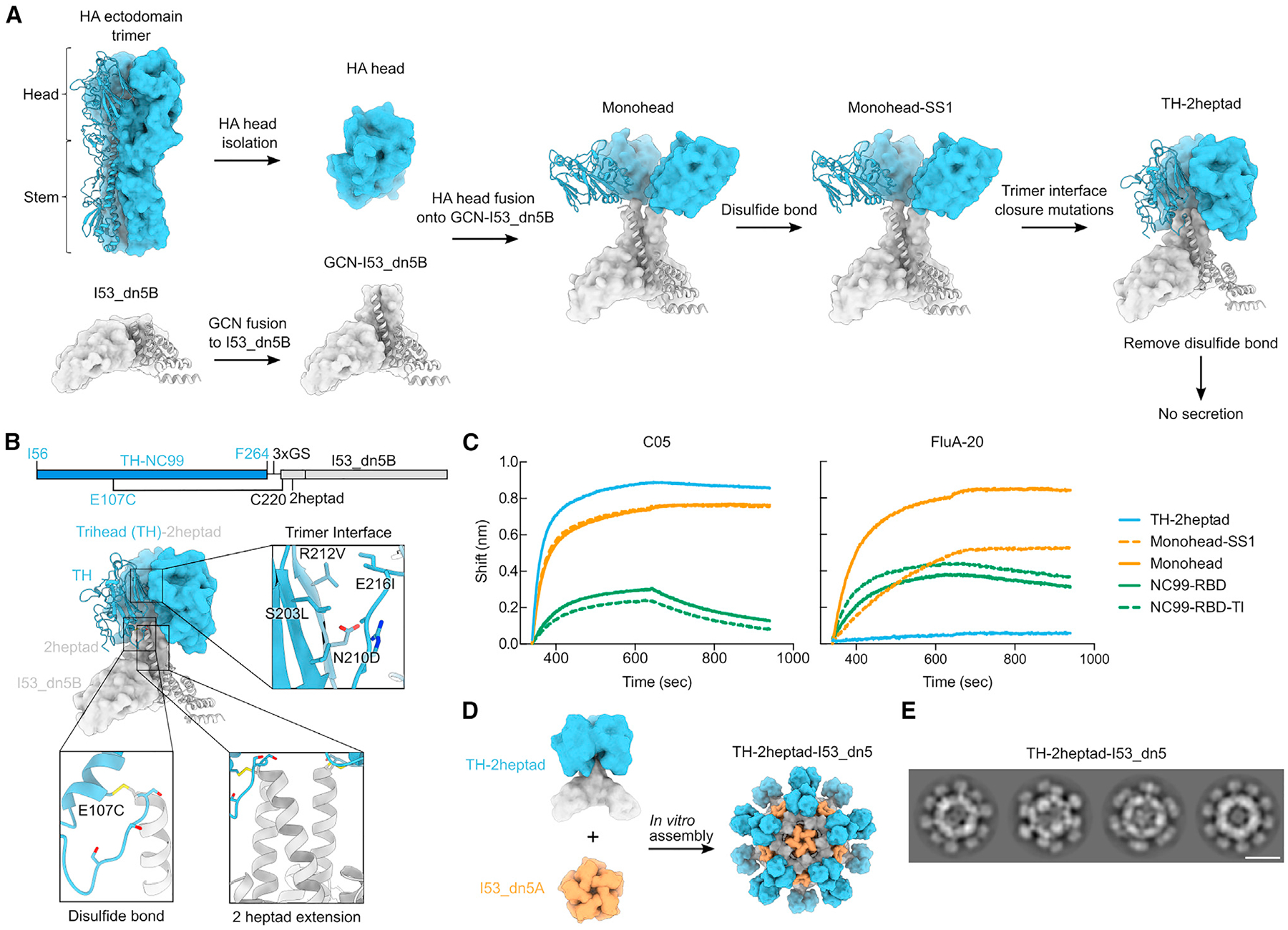
(A) Schematic of design process to make TH-2heptad construct. HA-derived segments are colored in blue, while segments derived from I53_dn5B and/or GCN4 are colored in gray.
(B) Gene diagram and model structure of TH-2heptad with closeups of the designed trimer interface, disulfide bond, and 2heptad extension domain. NC99 HA numbering is in blue, and trihead model numbering is in black.
(C) BLI of various NC99 RBD-based constructs against C05 and FluA-20.
(D) Schematic of in vitro assembly of the TH-2heptad-I53_dn5 nanoparticle.
(E) Cryo-EM 2D class averages of TH-2heptad-I53_dn5. Scale bar: 25 nm.
We therefore designed two additional structural elements to generate closed, rigidly displayed trimeric HA heads (referred to as “triheads” [THs]). First, we used Rosetta71 to identify a favorable intraprotomeric disulfide bond between the RBD and the N-terminal portion of the GCN4 bundle (Figures 1A and 1B). This monohead-SS1 construct was also confirmed to be a trimer by SEC but still bound to FluA-20 (Figure 1C). To further promote the closed trimeric state, we used Rosetta to optimize the hydrophobic interfaces between the RBDs while avoiding direct mutation of residues contacted by trimer-interface-directed antibodies27,29 (Figures 1A and 1B). A design containing the designed disulfide and four mutations, which we named TH-2heptad (S203L/N210D/R212V/E216I; Figure 1B), was successfully scaled up and purified by SEC, which confirmed its overall trimeric state (Figure S1A). In contrast to previous constructs, TH-2heptad did not bind to FluA-20, while a monomeric RBD containing the same trimer interface mutations, NC99-RBD-TI, bound as well as the wild-type NC99-RBD (Figure 1C). C05 binding was unaffected by the trimeric interface mutations, as expected. These results demonstrate that the trimer interface mutations help to stabilize the RBDs in a closed trimeric state, preventing FluA-20 access, but do not inherently disrupt the FluA-20 epitope.
We assembled TH nanoparticles by mixing purified TH-2heptad trimer with the complementary pentameric component I53_dn5A (Figure 1D)70 and purified them using SEC (Figures S1B and S1C). Cryo-electron microscopy (cryo-EM) 2D class averages of the nanoparticles revealed clear density for trimeric antigens on the surface of the nanoparticle (Figure 1E). The clarity with which the THs were resolved relative to the icosahedral nanoparticle scaffold sharply contrasts with previously described nanoparticle immunogens that used flexible genetic linkers62,72–77 and indicates rigid attachment to I53_dn5. Removal of the disulfide between E107C and C220 in the heptad domain abolished secretion, establishing that the combination of this disulfide and the trimer interface mutations was required for TH formation. Taken together, our BLI and cryo-EM data establish that TH-2heptad-I53_dn5 rigidly displays closed trimeric HA heads as intended.
Design and characterization of TH nanoparticles with varied antigen spacing
To enable investigation of the effects of antigen spacing on immunogenicity, we designed two new sequences that modified solely the heptad domain while retaining its rigidity. These were TH-1heptad, with one fewer heptad domain than TH-2heptad, and TH-6heptad, with four additional heptad domains that substantially lengthen the extendable region (Figure 2A). These constructs aimed to alter antigen-antigen spacing while keeping fixed the angle between the symmetry axes of neighboring antigens. We also designed a fourth “bobblehead” construct that introduced a glycine- and serine-based linker between the 2heptad domain and I53_dn5B to introduce flexibility between the antigens and I53_dn5 while maintaining trimeric closure of the RBDs. Finally, we included a control component, HA-I53_dn5B, which features the full NC99 HA ectodomain fused to I53_dn5B in the same manner as we previously reported.72 Each of these novel nanoparticle components were secreted from HEK293F cells and purified via IMAC and SEC (Figure S1A). All of the proteins, including the original monohead component, showed similar binding to the RBS-directed mAb C0522 and the lateral patch-targeting mAb Ab6649,35 demonstrating the preservation of these epitopes across all designs (Figure 2B). By contrast, FluA-20 binding was high only for the monohead, with very slight binding to TH-6heptad and HA-I53_dn5B and no binding to any of the other constructs, indicating stable RBD closure. As described above for TH-2heptad, the novel components were assembled in vitro with the I53_dn5A pentamer to form nanoparticles and purified via SEC (Figure S1B). The SEC chromatograms, SDS-PAGE (Figure S1C), dynamic light scattering (DLS; Figure 2C), and negative-stain EM (nsEM; Figure 2D) all indicated that the nanoparticles formed the intended assemblies with high homogeneity. The expected differences in size and antigen spacing between the three TH nanoparticles were evident in the DLS and nsEM data, respectively. The lack of significant amounts of residual, unassembled component during SEC indicated efficient assembly for all TH nanoparticles, bobblehead-I53_dn5, and HA-I53_dn5, whereas the monohead nanoparticle assembled less efficiently (Figure S1B; peak at ~16 mL). All TH nanoparticles showed melting temperatures that were similar to HA-I53_dn5 but higher than monohead-I53_dn5 (Figure S1D), suggesting that conformational stabilization of the RBDs in a trimeric state also reinforces their thermal stability.
Figure 2. Design and characterization of trihead nanoparticles with varied antigen spacing.
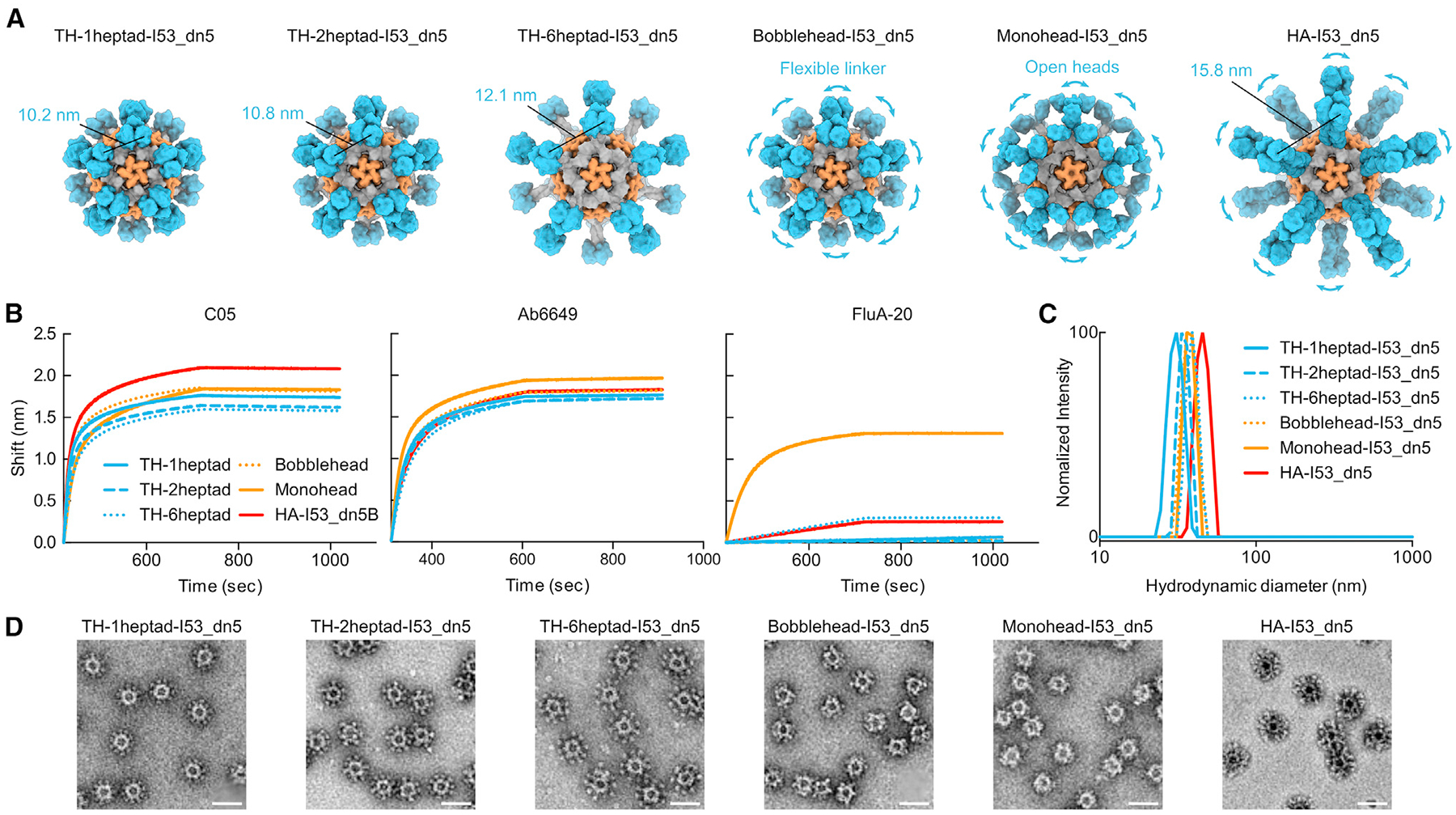
(A) Models of trihead nanoparticles with various extension domains, monohead-I53_dn5, and HA-I53_dn5. The models are colored blue, gray, and orange, respectively, for HA-derived segments, I53_dn5B/GCN4, and I53_dn5A.
(B) BLI of trihead nanoparticle extension series components against C05, FluA-20, and Ab6649.
(C) DLS of trihead nanoparticle extension series.
(D) nsEM micrographs of trihead nanoparticle extension series. Scale bars: 50 nm.
Cryo-EM of TH nanoparticle extension series
The structural differences among the TH, bobblehead, and monohead nanoparticles were examined using 3D reconstructions from either cryo-EM or nsEM. Cryo-EM structures were obtained for TH-1heptad-I53_dn5, TH-2heptad-I53_dn5, and TH-6heptad-I53_dn5 at respective resolutions of 4.1, 6.8, and 4.0Å (Figures 3A and S2A–S2C; Table S2). Compared to previous cryo-EM structures of trimeric antigen-nanoparticle fusions,62,72,73 the antigen density in this study was much more clearly defined, displaying the expected shape for three RBDs in the closed trimeric conformation symmetrically matched to the 3-fold symmetry axes of I53_dn5. The density observed between the RBDs and I53_dn5 was consistent with the heptad domain of each nanoparticle, with distinct lengths observed. However, the local resolution of the TH antigens was generally lower than that of I53_dn5, indicating some degree of flexibility in the connection between the antigens and the nanoparticle scaffold (Figure S2D). In contrast, 3D nsEM reconstructions for the bobblehead and monohead nanoparticles did not show clear density for the displayed antigens (Figures 3A and S2E), despite the presence of the stabilizing trimer interface mutations in bobblehead that eliminated binding to FluA-20 (Figure 2B). Consistent with their design, the monohead nanoparticle reconstruction revealed density related to its heptad domain rigidly attached to I53_dn5, while the reconstruction of the bobblehead nanoparticle, which contained an additional flexible linker between its heptad domain and I53_dn5, did not show such density.
Figure 3. Cryo-EM of trihead nanoparticle extension series.
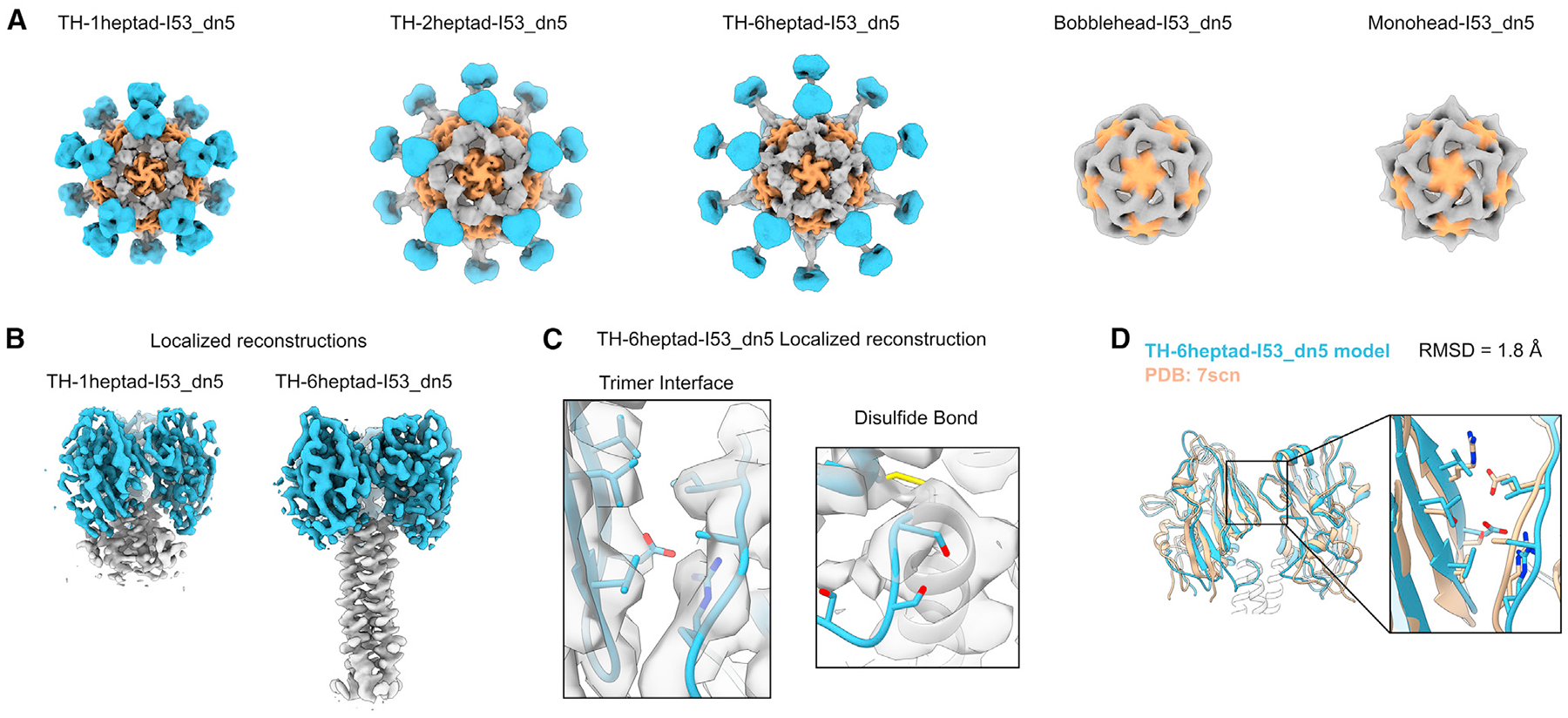
(A) Cryo-EM reconstructions of trihead nanoparticles and nsEM reconstructions of bobblehead and monohead nanoparticles. The densities are colored blue, gray, and orange, respectively, for HA-derived segments, I53_dn5B/GCN4, and I53_dn5A.
(B) Localized reconstructions of 1heptad and 6heptad triheads as displayed on I53_dn5 nanoparticles.
(C) Closeups of density with the corresponding built model from the TH-6heptad-I53_dn5 localized reconstruction for the engineered trimer interface and disulfide bond. Side chains for D210, R220, S266, S268, and S270 are only approximately displayed and are truncated in deposited maps.
(D) Superimposition of the TH-6heptad-I53_dn5 cryo-EM model with the NC99 HA crystal structure, with a blowup of the trimer interface where key interacting residues are displayed as sticks.
To gain a deeper understanding of the structures of the TH antigens, localized reconstructions of the RBD and heptad domains of TH-1heptad-I53_dn5 and TH-6heptad-I53_dn5 were generated with resolutions of 3.7 and 3.9Å, respectively (Figures 3B, S2A, and S2F). These reconstructions indicated rigid connections between the RBDs and the heptad domains, suggesting that the minor flexibility observed between the RBDs and the underlying nanoparticle likely derives from the heptad domains and their connections to I53_dn5 or that it is spread out across these domains. The atomic models of TH-1heptad and TH-6heptad antigens generated from these localized reconstructions confirmed the desired structural effects from the designed stabilizing features. Specifically, the inter-RBD interface showed hydrophobic packing between the designed mutations, and density consistent with disulfide formation was observed between E107C and GCN4, with less clear density where the flexible genetic linker was expected between each RBD and heptad domain (Figure 3C). The backbone atoms from all three TH-6heptad RBDs were structurally aligned with the RBDs from a cryo-EM structure of the NC99 trimeric ectodomain (PDB: 7SCN), revealing close similarity between the structures (1.8Å Cα root-mean-square deviation [RMSD]; Figure 1D). These detailed structural analyses complement our biophysical and antigenic data to confirm that the nanoparticle immunogens exhibit both of the target structural characteristics: stabilization of the RBD antigen in a native-like closed trimeric state and a rigid connection to the I53_dn5 nanoparticle that allows for variable antigen spacing.
Antibody responses in mice immunized with TH nanoparticle extension series
We next evaluated the immunogenicity of our TH nanoparticle extension series in mice. In a first study, groups of 10 BALB/c mice were immunized with AddaVax-adjuvanted protein at weeks 0, 4, and 8 (Figure S3A). Vaccine-matched NC99 HA-binding titers in week 10 sera were comparably high across all groups, with nearly all samples surpassing the upper limit of detection of the assay (Figure S3B). However, some differences in microneutralization titers were observed, with TH-1heptad-I53_dn5 and HA-I53_dn5 eliciting equivalent levels of neutralizing activity that were significantly higher than TH-6heptad-I53_dn5 and bobblehead-I53_dn5 (Figure S3C). Neutralizing activity within the TH extension series trended toward decreasing with increasing heptad region length. To expand the dynamic range of our serological readouts, we performed a similar study using unadjuvanted immunogens (Figure 4A). Analyzing immune sera from week 10 showed that TH-1heptad-I53_dn5 elicited significantly higher NC99 HAI titers than all other groups, including 5.3-fold higher titers than TH-6heptad-I53_dn5 and 5.2-fold higher titers than HA-I53_dn5 (Figure 4B). The pattern of microneutralization titers across the groups in this study trended similarly to the adjuvanted study, with the TH-1heptad-I53_dn5 and HA-I53_dn5 sera again having the highest neutralizing activity (Figure 4C). Lastly, in a second adjuvanted study, we analyzed binding titers in week 10 sera against three vaccine-mismatched H1 HAs (Figure 4D and S3D). The highest cross-reactive antibody titers were induced by monohead-I53_dn5 and HA-I53_dn5, possibly due to antibodies against conserved epitopes in the trimer interface and non-RBD domains, respectively. However, only these two nanoparticle immunogens elicited higher levels of cross-reactive antibodies than TH-1heptad-I53_dn5, which in turn elicited higher vaccine-mismatched binding titers than all of the other TH nanoparticles, though not always with statistical significance. Taken together, these results demonstrate a consistent trend toward increased serum antibody potency and breadth with decreased TH antigen spacing, with the TH-1heptad nanoparticle eliciting significantly more potent antibody responses than the other TH and monohead immunogens. Furthermore, the high levels of neutralizing activity and cross-reactive antibodies elicited by HA-I53_dn5 clearly indicate that deletion of domains outside the RBD can eliminate epitopes that aid in eliciting neutralizing and cross-reactive antibodies. Nevertheless, the dense display of closed trimeric RBDs in TH-1heptad-I53_dn5 provides an alternative approach to eliciting potent functional antibody responses, most notably through induction of higher levels of antibodies with HAI activity.
Figure 4. Antibody responses in mice immunized with trihead nanoparticle extension series.
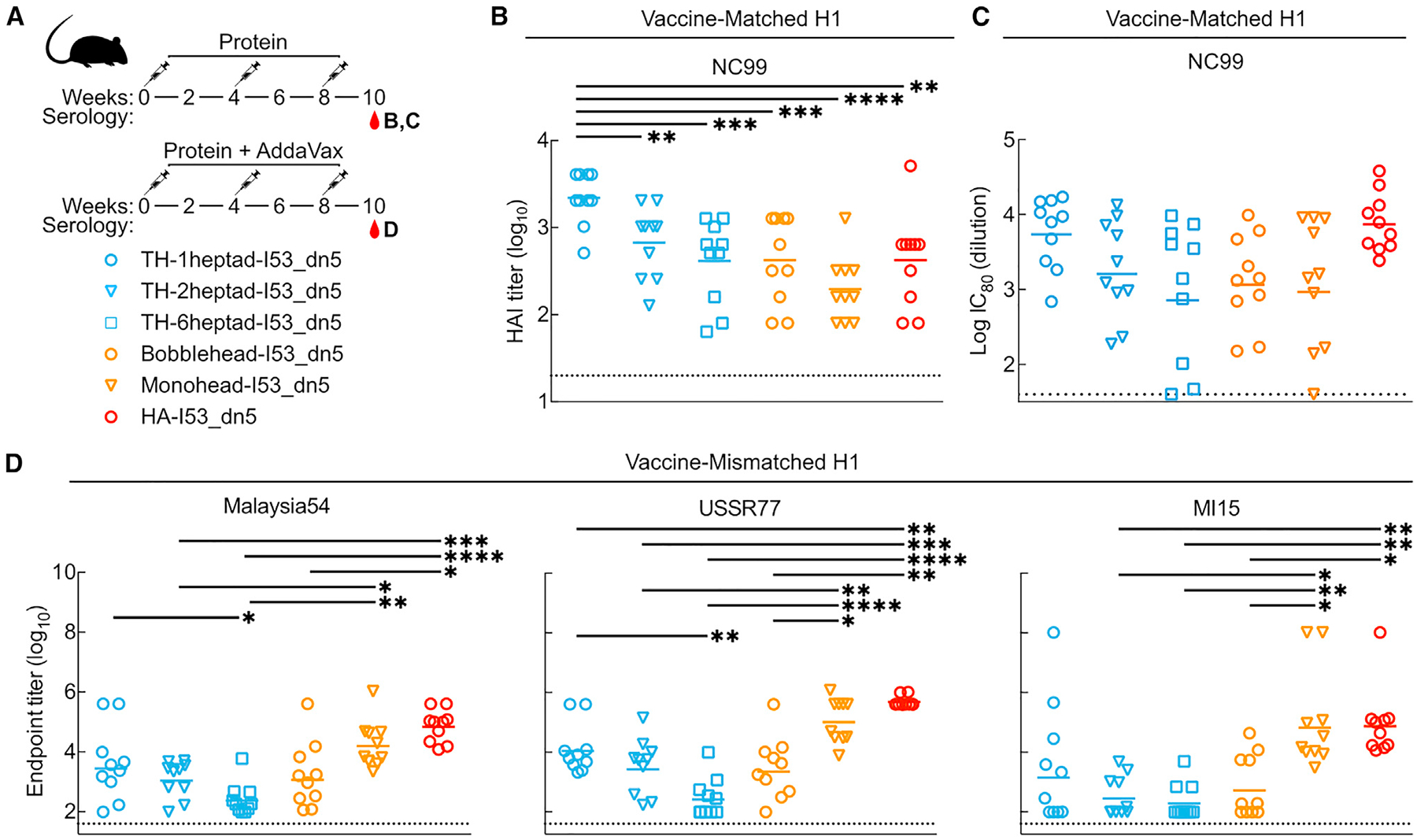
(A) Mouse immunization schedule and groups for trihead nanoparticle extension series that were either unadjuvanted or adjuvanted.
(B) Vaccine-matched HAI titers in unadjuvanted immune sera.
(C) Vaccine-matched microneutralization titers in unadjuvanted immune sera.
(D) Vaccine-mismatched H1 ELISA binding titers in adjuvanted immune sera.
Each symbol represents an individual animal, and the geometric mean of each group is indicated by the bar (n = 9–10 mice/group). Statistical significance was determined using one-way ANOVA with Tukey’s multiple comparisons test; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Epitope mapping of vaccine-elicited antibodies
To further understand how antigen conformation, spacing, and flexibility influence antibody responses against the RBD, we generated NC99-based HA probes featuring knockout mutations in specific epitopes to map the epitope specificities of vaccine-elicited antibodies. One set of probes comprised the trimeric HA ectodomain fused to foldon (referred to as NC99), with or without epitope knockout mutations. One RBS knockout probe was designed by introducing a single L194W mutation into the NC99 probe, replacing a highly conserved leucine residue in the center of the RBS with a bulkier tryptophan residue (Figure 5A). A second RBS knockout probe introduced an N-linked glycan within the RBS via T155N and K157T mutations. We expected the single L194W mutation to be more sensitive to antibodies that bind directly within the RBS pocket and that the bulky engineered glycan at residue 155 would interfere with binding to both RBS-directed and RBS-proximal antibodies. We also introduced the trimer interface mutations used in the TH antigen into the NC99 ectodomain trimer to maximally stabilize HA in the closed state (NC99-CS), as HA ectodomains are thought to breathe and transiently expose epitopes at the trimer interface.27,29,36,40 Finally, the wild-type monomeric NC99 RBD was also produced to allow detection of antibody binding anywhere on the HA head, including the trimer interface. Antigenic characterization of these probes was first performed using BLI. All of the probes bound to the lateral patch-targeting mAb Ab6649,35 indicating proper folding of the RBD (Figure S4A). As expected, all ectodomain trimer probes also bound to the anti-stem mAb CR9114,32 whereas the NC99-RBD did not. FluA-20 robustly bound the NC99-RBD, less efficiently bound NC99 and NC99-T155N/K157T, and did not bind NC99-L194W or NC99-CS, confirming that the trimer interface epitope is accessible in NC99-RBD but not NC99-CS. C05 bound NC99, NC99-CS, and NC99-RBD but did not bind either NC99-L194W or NC99-T155N/K157T, indicating that these modifications to the RBS prevent RBS-directed antibody binding as intended. All four trimeric ectodomain probes appeared mostly as small elongated rods by nsEM (Figure S4B), consistent with side views of HA ectodomain trimers. Some rosette formation was observed in all samples, especially NC99-T155N/K157T, as frequently occurs with recombinant HA trimers.78,79 Taken together, these results demonstrate that each probe possesses the desired structure and antigenicity.
Figure 5. Epitope mapping of vaccine-elicited antibodies.
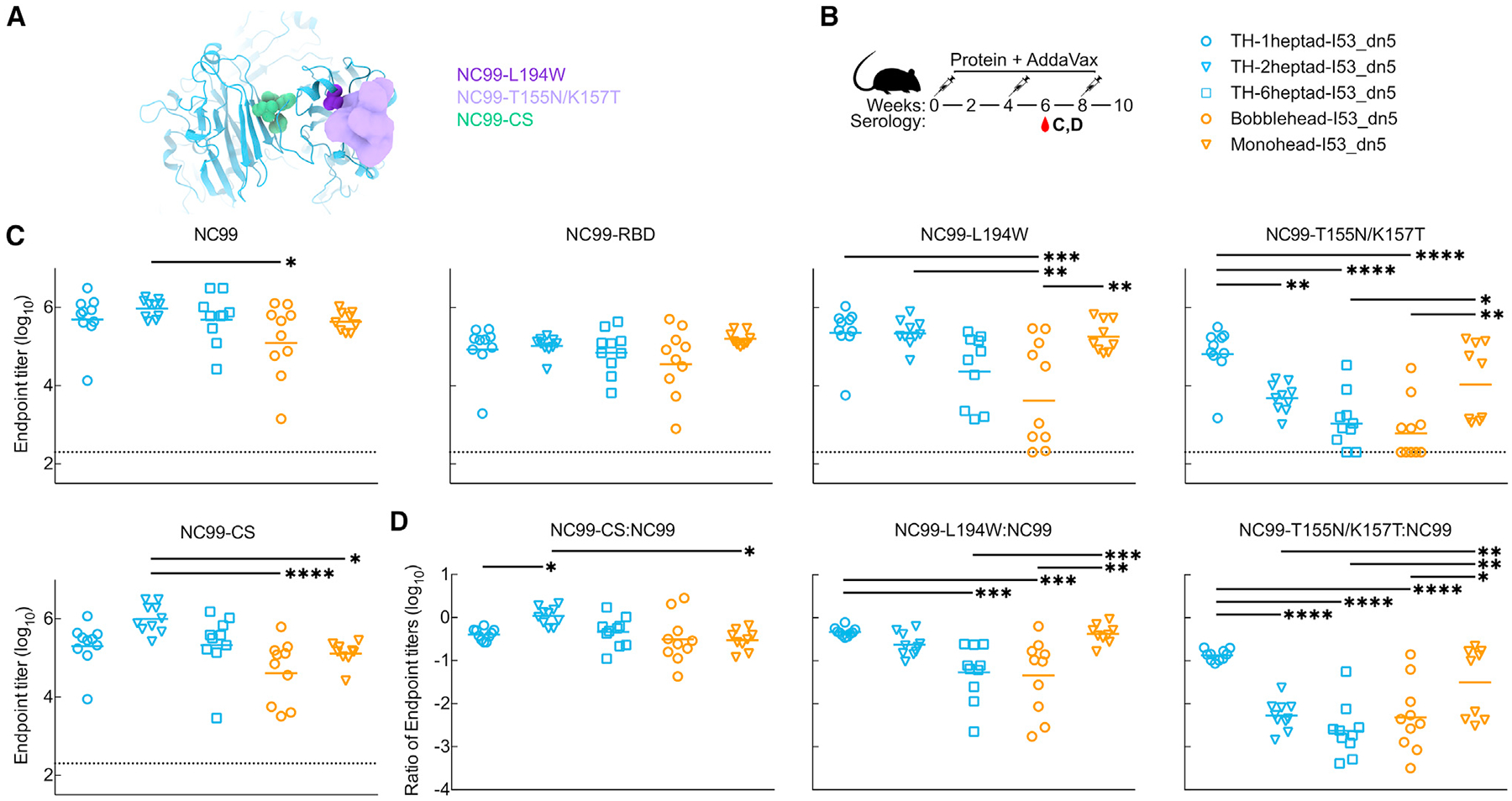
(A) Structural schematic of NC99 probes. Sphere representations show mutated residues. For NC99-T155N/K157T, the density of a modeled glycan is shown.
(B) Mouse immunization schedule and groups for adjuvanted trihead nanoparticle extension series.
(C) ELISA binding titers against various NC99 probes in week 6 adjuvanted immune sera.
(D) Ratios of ELISA binding titers of mutated NC99 probes to NC99 in (B).
Each symbol represents an individual animal, and the geometric mean of each group is indicated by the bar (n = 9–10 mice/group). Statistical significance was determined using one-way ANOVA with Tukey’s multiple comparisons test; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
These NC99 probes were then used to roughly estimate the epitope specificities of serum antibodies elicited by each immunogen, while the HA-I53_dn5 immunogen was not analyzed due to its inclusion of additional antigenic domains outside of the RBD. We used ELISA to measure the levels of antibodies against each probe in week 6 sera from our first adjuvanted study (Figures 5B and S3). All of the immunogens elicited similarly high binding titers against NC99 with the exception of bobblehead-I53_dn5, which was slightly lower (Figures 5C, S5A, and S5B). The same trend was observed in the NC99-RBD binding titers. Interestingly, the pattern of titers against the NC99-L194W and NC99-T155N/K157T RBS knockout probes mirrored the HAI and neutralization data (Figures 4B, 4C, and S3C) in that the titers elicited by the TH nanoparticle immunogens decreased with increasing heptad region length, while bobblehead-I53_dn5 elicited low and variable levels of antibodies against these probes. This pattern suggests that TH-1heptad elicited higher levels of antibodies directed at epitopes outside the RBS and its periphery, while the other immunogens elicited a higher proportion of responses targeting these epitopes. Ratiometric analysis of the binding of each serum sample to the two RBS knockout probes compared to NC99 is consistent with this suggestion (Figure 5D). It is also possible that differences in antibody affinity or avidity elicited by each immunogen could contribute to differences between these binding data and our HAI and neutralization data. Sera from mice that received monohead-I53_dn5 also appeared relatively insensitive to the RBS knockout mutations, although a subset of the sera showed substantially less binding to NC99-T155N/K157T (Figures 5C and 5D). Interestingly, these sera did not show significantly reduced binding to the NC99-CS probe, in which the TH interface mutations reduce exposure of the FluA-20 epitope (Figures 5C and 5D). Nevertheless, the insensitivity to the RBS knockout mutations and the cross-reactive antibody binding data in Figure 4D suggest that antibody responses against the trimer interface are elicited by monohead-I53_dn5. This conclusion is supported by our accompanying article in this issue of Cell Reports,80 which shows that hyperglycosylation of monohead-I53_dn5 can further increase the magnitude of the antibody response against the trimer interface relative to other epitopes on the RBD. In all, our data support the hypothesis that antigen display geometry can shape the magnitude and epitope specificities of vaccine-elicited antibodies, with TH-1heptad_I53_dn5 eliciting potent receptor-blocking antibody responses that nevertheless target a wider variety of epitopes compared to similar immunogens that vary only with respect to antigen display geometry.
DISCUSSION
Here, we designed a native-like trimeric HA RBD antigen that can be rigidly displayed on a protein nanoparticle scaffold and explored the effects of antigen display geometry on vaccine-elicited antibody responses. Combining domain-based antigens with nanoparticle display is an established and powerful strategy for enhancing the potency and breadth of antibody responses against epitopes that are targeted by potently neutralizing or protective antibodies. Domain-based antigens are small and often monomeric, making nanoparticle display particularly advantageous: in addition to improving their interactions with B cells,56,59–61,81 it also enhances their trafficking and localization in vivo by virtue of the increased size of the immunogen and, in some cases, the dense display of N-linked glycans.82–84 The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike RBD is a prominent example of a small monomeric antigen whose immunogenicity is greatly enhanced by multivalent display on protein nanoparticles,76,85,86 and the recent licensure of an RBD nanoparticle vaccine for SARS-CoV-2 has clinically validated this approach.87,88 Additional examples of nanoparticle display of domain-based antigens focusing responses on key neutralizing epitopes include Epstein-Barr virus gp35089 and human cytomegalovirus glycoprotein B.90 The strategy has also been combined with antigen stabilization,86,91,92 germline targeting,93–95 and mosaic nanoparticle display34,85,96,97 to further increase the potency and breadth of vaccine-elicited antibodies.
Displaying the influenza HA RBD in its native-like trimeric conformation required substantial structure-based antigen design. Our strategy was similar to those used to stabilize HA stem and coronavirus spike S2 antigens by deleting undesired domains and then repairing the resulting conformational changes or instabilities with stabilizing mutations or topological rearrangements.69,98–101 We focused on restoring the native-like closed trimeric arrangement of the displayed RBDs by strengthening the interactions between protomers, an approach that has been used to stabilize other oligomeric antigens in native-like conformations.69,102–110 Trimer closure required both computationally designed hydrophobic interactions at the trimer interface and a rigid, disulfide-mediated connection to an underlying trimerization domain. In our accompanying article in this issue of Cell Reports, we demonstrate that this strategy can be generalized to multiple H1 HAs and combined with additional antigen engineering to further refine vaccine-elicited antibody responses.80 These two features combine to establish THs as a robust and versatile antigen platform for influenza vaccine design. In the future, similar design approaches may be useful for stabilizing other oligomeric-domain-based antigens in native-like conformations.
In contrast to previous studies that aimed to focus responses on conserved subdominant epitopes by domain deletion69,98,99 or by reorienting antigens relative to particulate surfaces,111,112 our goal was to investigate how precise alterations to display geometry could influence antibody responses to a complex, domain-based antigen. To achieve this, we needed a system where display geometry could be modulated without affecting other vaccine characteristics that could impact antibody responses. Such a system requires a connection to the underlying nanoparticle that is both rigid and extendable. Previous work showed that DARPin proteins can be used as rigid adaptors between protein nanostructures and smaller monomeric proteins to facilitate structure determination by cryo-EM but did not allow for precise alteration of display geometry.113,114 We developed a nanoparticle component featuring an extendable N-terminal three-helix coiled coil and designed a rigid attachment between the TH antigen and the coiled coil that, to our knowledge, provides the most rigid and tunable connection between an antigen and protein nanoparticle scaffold to date. Although DNA origami has been used to precisely probe the effects of antigen organization on B cell activation in vitro,59,60 reports of the use of DNA origami nanoparticle vaccines in vivo are only beginning to appear.115,116 Elegant work displaying engineered HIV outer domain antigens on a series of protein nanoparticle scaffolds with different symmetries and valencies showed that responses to vaccination depend strongly on these structural factors and are driven by interactions with B cells.61 Our findings that altering antigen spacing alone can affect the HAI, microneutralization, cross-reactivity, and epitope specificity of vaccine-elicited antibodies further support that antigen organization is an important determinant of immunogenicity and establish that it can be optimized through structure-based antigen design.
Limitations of the study
There are several limitations to our study that present opportunities for future research. First, our serological data do not allow us to identify the mechanistic basis for the effects of antigen spacing on B cell recognition and antibody responses. Additionally, the icosahedral symmetry of I53_dn5 results in various geometric relationships between the displayed THs (e.g., between neighboring antigens across the icosahedral 2-fold or 5-fold symmetry axes), and it is unclear which relationships are most important for interactions with antigen-specific B cells. The forces experienced by immunogens during interactions with B cells are also poorly understood, and it is plausible that subtle conformational changes to the immunogen could occur during these interactions that deviate from our structural data. Furthermore, additional studies will be required to determine whether structural correlates of immunogenicity generalize across antigens since each antigen has unique structural properties. Finally, there are many factors beyond direct immunogen-B cell interactions that could affect the immunogenicity of our nanoparticle immunogen series in vivo, including antigen trafficking,82–84 conformational stability,117,118 and physical integrity,119 among others. To address these limitations, future studies will need to evaluate additional series of immunogens that systematically alter specific immunogen properties, an endeavor that will be facilitated by powerful new machine learning methods for protein structure prediction and design.120–123 Corresponding advances in in vitro studies of antigen-B cell interactions124 and animal models of vaccination61,125–128 are providing increasingly precise measurements of immunogen performance in vitro and in vivo. Combining these technologies to define structural correlates of immunogenicity could be a powerful route to improved vaccines against influenza and many other pathogens.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Neil King (neilking@uw.edu).
Materials availability
All unique and stable materials generated in this study are available from the lead contact under a Materials Transfer Agreement.
Data and code availability
Structural data generated from this study were deposited in public data repositories and the accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| C05 | Produced in house (Ekiert et al.)22 | N/A |
| FluA-20 | Produced in house (Bangaru et al.)29 | N/A |
| Ab6649 | Produced in house (Raymond et al.)35 | N/A |
| Goat anti-human IgG, HRP | SouthernBiotech | Cat# 2040–05; RRID:AB_2795644 |
| Goat anti-mouse IgG, HRP | SouthernBiotech | Cat# 1080–05; RRID:AB_2734756 |
| Bacterial and virus strains | ||
| A/New Caledonia/20/1999 R3ΔPB1 virus | Produced in house | N/A |
| Influenza reporter viruses | Produced in house (Creanga et al.)147 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| AddaVax adjuvant | Cat# vac-adx-10 | |
| TMB | SeraCare | Cat# 5120–0083 |
| Polyethylenimine (PEI) HCl MAX, Linear, Mw 40,000 | Polysciences | Cat# 24765–1 |
| PNGase F | NEB | Cat# P0705 |
| RDE II | VWR | Cat# 10753–482 |
| Deposited data | ||
| Trihead-1heptad-I53_dn5 | EMDataBank | EMD: 42481 |
| Localized reconstruction of Trihead-1heptad-I53_dn5 | PDB, EMDataBank | PDB ID: 8UR5, EMD: 42482 |
| Trihead-2heptad-I53_dn5 | EMDataBank | EMD: 42483 |
| Trihead-6heptad-I53_dn5 | EMDataBank | EMD: 42485 |
| Localized reconstruction of Trihead-6heptad-I53_dn5 | PDB, EMDataBank | PDB ID: 8UR7, EMD:42486 |
| Experimental models: Cell lines | ||
| Expi293F | ThermoFisher | Cat# A14527 |
| MDCK-SIAT-PB1 | Produced in house (Creanga et al.)147 | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c mice | Jackson Laboratory | Cat# 000651 |
| Recombinant DNA | ||
| See Table S1 for amino acid sequences | ||
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Leginon | (Suloway et al.)132 | N/A |
| CryoSparc | (Punjanietal.)134 | https://cryosparc.com/ |
| UCSF Chimera | (Pettersen et al.)141 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX | (Goddard et al.)145 | https://www.rbvi.ucsf.edu/chimerax/ |
| PyMol | Schrodinger, LLC | https://pymol.org/2/ |
| Unicorn 7.0 | GE Healthcare | http://www.gelifesciences.com/ |
| Relion | (Zivanov et al.)135 | N/A |
| Other | ||
| 300 mesh grids | Ted Pella | Cat# 01843-F |
| UltrAufoil 2/2, 200 mesh grids | Electron Microscopy Sciences | |
| Filter paper | Cytiva | Cat# 1004047 |
| Uranyl formate | SPI Chem | Cat# 02545-AA |
| Superdex 200 Increase SEC column | Cytiva | Cat# 28-9909-44 |
| Superose 6 Increase SEC column | Cytiva | Cat# 29091596 |
| Excel resin | Cytiva | Cat# 17371203 |
| Isoflurane | USP Patterson | Cat# 07-893-1389 |
| EndoSafe LAL Test Cartridges | Charles River Labs | Cat# PTS20005F |
| Lemo21(DE3) | New England BioLabs | Cat# C2528J |
| MAX Efficiency™ DH5α Competent Cells | Sigma-Aldrich | Cat# I6758 |
| Isopropyl-B-D-thiogalactoside (IPTG) | Thermo Fisher | Cat# 18258012 |
| Kanamycin Sulfate | Sigma-Aldrich | Cat# K1876 |
| HisTrap HP | Cytiva | Cat#17-5247-01 |
| Octet Protein A Biosensors | Sartorius | Cat# 18-5010 |
| Turkey Red Blood Cells | Lampire | Cat# # 7209403 |
| Plasmid Plus Maxi Kit | Qiagen | Cat# 12963 |
| Trypsin, TPCK Treated | Thermo Fisher | Cat# 20233 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Expi293F cells are derived from the HEK293F cell line (Life Technologies). Expi293F cells were grown in Expi293 Expression Medium (Life Technologies), cultured at 36.5°C with 8% CO2 and shaking at 150 rpm.
Mice
Female BALB/c mice 4–6 weeks old were obtained from Jackson Laboratory, Bar Harbor, Maine, and maintained at the Vaccine Research Center (VRC), NIH. All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the VRC, NIAID, NIH. All animals were housed and cared for in compliance with all federal regulations, NIH guidelines, AAALAC, and IACUC approval. Only female mice were used to enable evaluation of the immunogens in a consistent model immune system. As a result, we did not evaluate sex as a biological variable.
METHOD DETAILS
Structure-based design
All design and modeling was performed using Rosetta and PyMOL. Briefly, modeling of linker regions was performed using RosettaRemodel.129 Disulfide design between the HA RBD and the GCN4-based rigid linker region (PDB: 1GCM) was assessed using the Disulfidize mover after symmetrically superimposing HA RBDs (PDB: 5UG0) on the rigid linker region based on the native orientation of HA RBDs to the apical region of the HA2 domain, which is structurally similar to GCN4. To determine stabilizing mutations at the HA RBD interface, Rosetta design was performed on residues 203, 210, 212 and 216 of PDB 5UG0 using resfile-based design methods described previously91,102 with C3-symmetry imposed to model the closed state of the HA heads. Mutations were then transferred to the NC99 sequence to complete the design sequence.
Gene expression and protein purification
All HA constructs used in this study contained the Y98F mutation to reduce aggregation,130 were codon-optimized for human cell expression, and cloned into the CMV/R vector131 by Genscript with a C-terminal hexahistidine affinity tag. PEI MAX was used for transient transfection of HEK293F cells. After four days, mammalian cell supernatants were clarified via centrifugation and filtration. Monohead and trihead components and HA foldons were all purified using IMAC. 1 mL of Ni2+-sepharose Excel or Talon resin was added per 100 mL clarified supernatant along with 5 mL of 1 M Tris, pH 8.0 and 7 mL of 5 M NaCl and left to batch bind while shaking at room temperature for 30 min. Resin was then collected in a gravity column, washed with 5 column volumes of 50 mM Tris, pH 8.0, 500 mM NaCl, 20 mM imidazole, and protein was eluted using 50 mM Tris, pH 8.0, 500 mM NaCl, 300 mM imidazole. Further component purification was done using SEC on a Superdex 200 Increase 10/300 gel filtration column equilibrated in 25 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol.
Expression and purification of the I53_dn5A pentamer component from E. coli was carried out by IPTG induction and IMAC followed by SEC on a Superdex 200 Increase 10/300 gel filtration column, respectively72, including endotoxin removal through the inclusion of CHAPS detergent in purification buffers, using the stabilized I53_dn5A.1 sequence.70 Assembly of all nanoparticles was carried out by mixing HA-I53_dn5B-based components and pentameric I53_dn5A together in vitro at a 1:1 M ratio at 15–40 μM final concentrations. Nanoparticles were left to assemble for 30 min at room temperature with rocking. Nanoparticles were then purified using SEC on a Superose 6 Increase 10/300 gel filtration column equilibrated in 25 mM Tris, pH 8.0, 150 mM NaCl, 5% glycerol.
Following purification, nanoparticle quality and concentration was first measured by UV-vis spectroscopy. Nanoparticle polydispersity and purity was then assessed using SDS-PAGE, DLS, and nsEM. Finally, endotoxin levels were measured using the LAL assay, with all immunogens used in animal studies containing less than 100 EU/mg in the final dose. Final immunogens were flash-frozen using liquid nitrogen and stored at −80°C.
Bio-layer interferometry (BLI)
BLI was carried out using an Octet Red 96 system, at 25°C with 1000 rpm shaking. Anti-HA antibodies were diluted in kinetics buffer (PBS with 0.5% serum bovine albumin and 0.01% Tween) to a final concentration of 10 μg/mL before loading onto protein A biosensors (Sartorius) for 200 s. Proteins were diluted to 400–500 nM in kinetics buffer and their association was measured for 200 s, followed by dissociation for 200 s in kinetics buffer alone.
Dynamic light scattering
DLS was carried out on an UNcle (UNchained Labs) at 25°C. 10 acquisitions of 5 s each were acquired for each spectrum. Protein concentration (ranging from 0.1–1 mg/mL) and buffer conditions were accounted for in the software.
Negative stain EM
Nanoparticle samples were diluted to 0.01 mg/mL immediately prior to adsorption to glow-discharged carbon-coated copper grids for about 60 s prior to a 2% uranyl formate staining. Micrographs were recorded using the Leginon132 on a 120 kV FEI Tecnai G2 Spirit with a Gatan Ultrascan 4000 4k × 4k CCD camera at 67,000× nominal magnification. The defocus ranged from −1.0 to −2.0 μm and the pixel size was 1.6Å.
Cryo-EM sample preparation, data collection, and data processing
Three microliters of 3 mg/mL TH-1heptad-I53_dn5, TH-2heptad-I53_dn5 or TH-6heptad-I53_dn5 nanoparticle samples were loaded onto freshly glow discharged R 2/2 UltrAuFoil grids, prior to plunge freezing using a Vitrobot Mark IV (ThermoFisher Scientific) with a blot force of 0 and 6 s blot time at 100% humidity and 22°C. For the TH-1heptad-I53_dn5 and TH-6heptad-I53_dn5 datasets, data were acquired on an FEI Glacios transmission electron microscope operated at 200 kV equipped with a Gatan K2 Summit direct detector. Automated data collection was carried out using Leginon132 at a nominal magnification of 36,000× with a pixel size of 1.16Å. The dose rate was adjusted to 8 counts/pixel/s, and each movie was acquired in counting mode fractionated in 50 frames of 200 ms. 1,274 and 1,647 micrographs were collected with a defocus range between −0.5 and −2.5 μm. For the TH-2heptad-I53_dn5 dataset, data were acquired using an FEI Titan Krios transmission electron microscope operated at 300 kV and equipped with a Gatan K2 direct detector and Gatan Quantum GIF energy filter, operated in zero-loss mode with a slit width of 20 eV. Automated data collection was carried out using Leginon132 at a nominal magnification of 130,000× with a pixel size of 0.525Å. 224 micrographs were collected with a defocus range comprised between −0.5 and −2.5 μm, respectively. The dose rate was adjusted to 15 counts/pixel/s, and each movie was acquired in super-resolution mode fractionated in 75 frames of 40 ms. Movie frame alignment, estimation of the microscope contrast-transfer function parameters, particle picking, and extraction were carried out using Warp.133 Two rounds of reference-free 2D classification were performed using CryoSPARC134 to select well-defined particle images. These selected particles were subjected to two rounds of 3D classification with 50 iterations each (angular sampling 7.5° for 25 iterations and 1.8° with local search for 25 iterations) using Relion135 with an initial model generated with ab-initio reconstruction in CryoSPARC.134 3D refinements with icosahedral symmetry were carried out using non-uniform refinement along with per-particle defocus refinement in CryoSPARC. Selected particle images were subjected to the Bayesian polishing procedure136 implemented in Relion 3.1 before performing another round of non-uniform refinement in cryoSPARC followed by per-particle defocus refinement and again non-uniform refinement. To further improve the density of the trimeric HA head, a localized reconstruction method was used.137 The designed asymmetric unit models were fitted to the icosahedral map and the locations of 20 HA RBD trimers in the nanoparticles were manually defined in UCSF Chimera by placing a marker in the center of each HA RBD trimer. A total of 1,452,820 sub-particles and 718,940 sub-particles were selected for TH-1heptad HA trihead and TH-6heptad HA trihead respectively using Scipion.138 The selected sub-particles were extracted in a box of 180 × 180 pixels from micrographs and subjected to one round of 3D classification using Relion.135 Particles belonging to classes with the best resolved trimeric HA trihead density were selected and then subjected to local refinement with C3 symmetry using CryoSPARC. Local resolution estimation and sharpening were carried out using CryoSPARC. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) of 0.143 criterion and Fourier shell correlation curves were corrected for the effects of soft masking by high-resolution noise substitution.139,140
Model building and refinement
UCSF Chimera141 and Coot142 were used to build atomic models that fit into the cryo-EM density maps. TH-1heptad and TH-6heptad HA trihead models were refined and relaxed using Rosetta using sharpened and unsharpened maps.143,144 Figure models were generated using ChimeraX.145
Animal experiments
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the VRC, NIAID, NIH. All animals were housed and cared for in accordance with local, state, federal, and institutional policies of NIH and American Association for Accreditation of Laboratory Animal Care. The space temperature in the rodent facility is set to 22°C ± 3°. The humidity is maintained between 30% and 70%. The automatic light cycle is a 12 h on/off photo-period.
Immunization
Mice were immunized intramuscularly with 22 μmol purified nanoparticle immunogen in the presence or absence of AddaVax (InvivoGen) at weeks 0, 4, and 8. Formulated nanoparticle immunogens in 50 μL were given into each hind leg. Serum samples were collected before and after each immunization and used for immunological assays. (Figures 4, 5, and S3).
ELISA
Antigen-specific IgG levels in immune sera were measured by ELISA. The plates were coated with 2 μg mL−1 of recombinant HA-foldon proteins and incubated at 4°C overnight. Plates were then blocked with PBS containing 5% skim milk at 37°C for 1 h. mAbs and immune sera were serially diluted in 4-fold steps and added to the wells for 1 h. Horseradish peroxidase (HRP)-conjugated anti-human (SouthernBiotech, Catalog 2040–05, used 1/5,000) or anti-mouse IgG (SouthernBiotech, Catalog 1080–05, used 1/5000) antibody was added and incubated at 37°C for 1 h. The wells were developed with 3,3′,5′,5-tetramethylbenzidine (TMB) substrate (KPL), and the reactions were stopped by adding 1 M H2SO4 before measuring absorbance at 450 nm with a Spectramax Paradigm plate reader (Molecular Devices). Sera from mice immunized with PBS or an irrelevant antigen (DS-Cav1-I53_dn5;70) were used as negative controls, and did not yield signal above background. Raw data were analyzed in GraphPad Prism and endpoint titers were recorded as the last serum dilution resulting in a 4-fold increase in absorbance above background.
HAI
Influenza antibody titers were detected using HAI (Hemagglutination Inhibition) with 2-fold serial dilutions of RDE-treated mouse serum.146 One part serum sample was mixed with three parts of diluted receptor destroying enzyme (RDE II; Denka Seiken) and treated at 37°C for 16–18 h followed by heat inactivation at 56°C for 40 min. RDE-treated serum was then serially diluted in a 96-well format with PBS (total final volume of 25 μL per dilution) and 25 μL of A/New Caledonia/20/1999 R3ΔPB1 virus (at a concentration of 8HA) was added to each well with mixing. After a 1-h incubation at room temperature, 50 μL of 0.5% washed turkey whole blood (Lampire, cat# 7209403) was added to each well. The samples were allowed to incubate an additional 30 min at room temperature, and the serum dilution that no longer successfully inhibited hemagglutination as seen by drip test was documented as the HAI titer for each sample.
Reporter-based microneutralization assay
Influenza A Reporter viruses were made from H1N1 viruses with a modified PB1 segment expressing the TdKatushka reporter gene (R3ΔPB1), rescued, and propagated in MDCK-SIAT-PB1 cells in the presence of TPCK-treated trypsin (1 μg mL−1, Sigma) at 37 °C.147 Virus stocks were stored at −80 °C and were titrated before use in the assay. Mouse sera were treated with receptor destroying enzyme (RDE II; Denka Seiken) and heat-inactivated before use in neutralization assays. 384-well plates (Grenier) were pre-seeded with 1.0×105 MDCK-SIAT1-PB1 cells and incubated overnight. Immune sera or monoclonal antibodies (FluA-20, Ab6649, C05) were serially diluted and incubated for 1 h at 37 °C with pre-titrated virus (A/New Caledonia/20/1999). Serum-virus mixtures were then transferred in quadruplicate onto the pre-seeded 384-well plates and incubated at 37 °C for 18–26 h. The number of fluorescent cells in each well was counted automatically using a Celigo image cytometer (Nexcelom Biosciences). IC80 values, defined as the serum dilution or antibody concentration that gives 80% reduction in virus-infected cells, were calculated from neutralization curves using a four-parameter nonlinear regression model.
Statistical analysis
Multi-group comparisons were performed using the Brown-Forsythe one-way ANOVA test and Dunnett’s T3 post hoc analysis in Prism 9 (GraphPad). Differences were considered significant when p values were less than 0.05.
Supplementary Material
Highlights.
Computational design of a closed trimeric HA RBD (“trihead”) antigen platform
Design of a rigid, extendable linker enables precise variation of antigen spacing
Decreased antigen spacing of triheads elicits responses with increased breadth and potency
Changes to antigen spacing alter epitope specificities of vaccine-elicited antibodies
ACKNOWLEDGMENTS
This work was funded by a generous gift from Open Philanthropy (N.P.K.); the Audacious Project at the Institutefor Protein Design (N.P.K.); the Defense Threat Reduction Agency (HDTRA1-18-1-0001 to N.P.K.); the National Institute of Allergy and Infectious Diseases (P01 AI167966 to D.V. and N.P.K.); the intramural research program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (M.K. and B.S.G.); the University of Washington Arnold and Mabel Beckman Cryo-EM Center; and the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001 (Y.T.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113552.
DECLARATION OF INTERESTS
N.P.K. is a co-founder, shareholder, paid consultant, and chair of the scientific advisory board of Icosavax, Inc. The King lab has received unrelated sponsored research agreements from Pfizer and GSK. D.E. is a shareholder of Icosavax, Inc. A.D., D.E., M.K., and N.P.K. are listed as co-inventors on patent applications filed by the University of Washington related to this work.
REFERENCES
- 1.Harrison SC (2015). Viral membrane fusion. Virology 479–480, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu NC, and Wilson IA (2020). Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb. Perspect. Med 10, a038778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F, and Palese P (2015). Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov 14, 167–182. [DOI] [PubMed] [Google Scholar]
- 4.Dugan HL, Guthmiller JJ, Arevalo P, Huang M, Chen Y-Q, Neu KE, Henry C, Zheng N-Y, Lan LY-L, Tepora ME, et al. (2020). Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med 12, eabd3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angeletti D, and Yewdell JW (2018). Understanding and Manipulating Viral Immunity: Antibody Immunodominance Enters Center Stage. Trends Immunol. 39, 549–561. [DOI] [PubMed] [Google Scholar]
- 6.Guthmiller JJ, Utset HA, and Wilson PC (2021). B Cell Responses against Influenza Viruses: Short-Lived Humoral Immunity against a Life-Long Threat. Viruses 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rimmelzwaan GF, and McElhaney JE (2008). Correlates of protection: novel generations of influenza vaccines. Vaccine 26 (Suppl 4), D41–D44. [DOI] [PubMed] [Google Scholar]
- 8.Knight M, Changrob S, Li L, and Wilson PC (2020). Imprinting, immunodominance, and other impediments to generating broad influenza immunity. Immunol. Rev 296, 191–204. [DOI] [PubMed] [Google Scholar]
- 9.Belongia EA, Simpson MD, King JP, Sundaram ME, Kelley NS, Osterholm MT, and McLean HQ (2016). Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect. Dis 16, 942–951. [DOI] [PubMed] [Google Scholar]
- 10.Ortiz JR, Bernstein DI, Hoft DF, Woods CW, McClain MT, Frey SE, Brady RC, Bryant C, Wegel A, Frenck RW, et al. (2023). A Multi-Center, Controlled Human Infection Study of Influenza A (H1N1) pdm09 in Healthy Adults. J. Infect. Dis 228, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunning AJ, DiazGranados CA, Voloshen T, Hu B, Landolfi VA, and Talbot HK (2016). Correlates of Protection against Influenza in the Elderly: Results from an Influenza Vaccine Efficacy Trial. Clin. Vaccine Immunol 23, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhard W, Yewdell J, Frankel ME, and Webster R (1981). Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290, 713–717. [DOI] [PubMed] [Google Scholar]
- 13.Caton AJ, Brownlee GG, Yewdell JW, and Gerhard W (1982). The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–427. [DOI] [PubMed] [Google Scholar]
- 14.Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. (2017). Defining B cell immunodominance to viruses. Nat. Immunol 18, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiley DC, Wilson IA, and Skehel JJ (1981). Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289, 373–378. [DOI] [PubMed] [Google Scholar]
- 16.Bajic G, and Harrison SC (2021). Antibodies That Engage the Hemagglutinin Receptor-Binding Site of Influenza B Viruses. ACS Infect. Dis 7, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zost SJ, Lee J, Gumina ME, Parkhouse K, Henry C, Wu NC, Lee C-CD, Wilson IA, Wilson PC, Bloom JD, and Hensley SE (2019). Identification of Antibodies Targeting the H3N2 Hemagglutinin Receptor Binding Site following Vaccination of Humans. Cell Rep. 29, 4460–4470.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr., and Wilson IA (2013). A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat. Struct. Mol. Biol 20, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong M, Lee PS, Hoffman RMB, Zhu X, Krause JC, Laursen NS, Yoon S-I, Song L, Tussey L, Crowe JE Jr., et al. (2013). Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol 87, 12471–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. (2013). Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. USA 110, 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 108, 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, et al. (2012). Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, and Wilson IA (2012). Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. USA 109, 17040–17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, and Wilson IA (2014). Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun 5, 3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Chen J, Zheng Q, Xue W, Zhang L, Rong R, Zhang S, Wang Q, Hong M, Zhang Y, et al. (2022). Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses. Nat. Commun 13, 5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, and Harrison SC (2018). Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 48, 174–184.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, et al. (2019). Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 177, 1124–1135.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zost SJ, Dong J, Gilchuk IM, Gilchuk P, Thornburg NJ, Bangaru S, Kose N, Finn JA, Bombardi R, Soto C, et al. (2021). Canonical features of human antibodies recognizing the influenza hemagglutinin trimer interface. J. Clin. Invest 131, e146791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, et al. (2019). A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 177, 1136–1152.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee C-H, Lavinder JJ, et al. (2016). Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat. Med 22, 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X, Guo Y-H, Jiang T, Wang Y-D, Chan K-H, Li X-F, Yu W, McBride R, Paulson JC, Yuen K-Y, et al. (2013). A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J. Virol 87, 12619–12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krause JC, Tumpey TM, Huffman CJ, McGraw PA, Pearce MB, Tsibane T, Hai R, Basler CF, and Crowe JE Jr. (2010). Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J. Virol 84, 3127–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. (2019). Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol 20, 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, and Harrison SC (2018). Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 115, 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner HL, Pallesen J, Lang S, Bangaru S, Urata S, Li S, Cottrell CA, Bowman CA, Crowe JE Jr., Wilson IA, and Ward AB (2019). Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PLoS Biol. 17, e3000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen JD, Ray S, and Ross TM (2018). Split inactivated COBRA vaccine elicits protective antibodies against H1N1 and H3N2 influenza viruses. PLoS One 13, e0204284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giles BM, and Ross TM (2011). A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29, 3043–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sautto GA, Kirchenbaum GA, and Ross TM (2018). Towards a universal influenza vaccine: different approaches for one goal. Virol. J 15, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, and Schmidt AG (2019). Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 25, 827–835.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thornlow DN, Macintyre AN, Oguin TH, Karlsson AB, Stover EL, Lynch HE, Sempowski GD, and Schmidt AG (2021). Altering the Immunogenicity of Hemagglutinin Immunogens by Hyperglycosylation and Disulfide Stabilization. Front. Immunol 12, 737973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajic G, Maron MJ, Caradonna TM, Tian M, Mermelstein A, Fera D, Kelsoe G, Kuraoka M, and Schmidt AG (2020). Structure-Guided Molecular Grafting of a Complex Broadly Neutralizing Viral Epitope. ACS Infect. Dis 6, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caradonna TM, Ronsard L, Yousif AS, Windsor IW, Hecht R, Bracamonte-Moreno T, Roffler AA, Maron MJ, Maurer DP, Feldman J, et al. (2022). An epitope-enriched immunogen expands responses to a conserved viral site. Cell Rep. 41, 111628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun W, Kirkpatrick E, Ermler M, Nachbagauer R, Broecker F, Krammer F, and Palese P (2019). Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol 93, e00333–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broecker F, Liu STH, Suntronwong N, Sun W, Bailey MJ, Nachbagauer R, Krammer F, and Palese P (2019). A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nachbagauer R, Feser J, Naficy A, Bernstein DI, Guptill J, Walter EB, Berlanda-Scorza F, Stadlbauer D, Wilson PC, Aydillo T, et al. (2021). A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med 27, 106–114. [DOI] [PubMed] [Google Scholar]
- 47.Krammer F, Pica N, Hai R, Margine I, and Palese P (2013). Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol 87, 6542–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu X, Han J, Sun W, Puente-Massaguer E, Yu W, Palese P, Krammer F, Ward AB, and Wilson IA (2022). Influenza chimeric hemagglutinin structures in complex with broadly protective antibodies to the stem and trimer interface. Proc. Natl. Acad. Sci. USA 119, e2200821119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irvine DJ, and Read BJ (2020). Shaping humoral immunity to vaccines through antigen-displaying nanoparticles. Curr. Opin. Immunol 65, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, and Subramaniam S (2013). Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc. Natl. Acad. Sci. USA 110, 4592–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein JS, and Bjorkman PJ (2010). Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6, e1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amitai A, Sangesland M, Barnes RM, Rohrer D, Lonberg N, Lingwood D, and Chakraborty AK (2020). Defining and Manipulating B Cell Immunodominance Hierarchies to Elicit Broadly Neutralizing Antibody Responses against Influenza Virus. Cell Syst. 11, 573–588.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiller J, and Chackerian B (2014). Why HIV virions have low numbers of envelope spikes: implications for vaccine development. PLoS Pathog. 10, e1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachmann MF, and Zinkernagel RM (1996). The influence of virus structure on antibody responses and virus serotype formation. Immunol. Today 17, 553–558. [DOI] [PubMed] [Google Scholar]
- 55.Bachmann MF, Kalinke U, Althage A, Freer G, Burkhart C, Roost H, Aguet M, Hengartner H, and Zinkernagel RM (1997). The role of antibody concentration and avidity in antiviral protection. Science 276, 2024–2027. [DOI] [PubMed] [Google Scholar]
- 56.Bachmann MF, and Jennings GT (2010). Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol 10, 787–796. [DOI] [PubMed] [Google Scholar]
- 57.Dintzis HM, Dintzis RZ, and Vogelstein B (1976). Molecular determinants of immunogenicity: the immunon model of immune response. Proc. Natl. Acad. Sci. USA 73, 3671–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chackerian B, Lenz P, Lowy DR, and Schiller JT (2002). Determinants of autoantibody induction by conjugated papillomavirus virus-like particles. J. Immunol 169, 6120–6126. [DOI] [PubMed] [Google Scholar]
- 59.Shaw A, Hoffecker IT, Smyrlaki I, Rosa J, Grevys A, Bratlie D, Sandlie I, Michaelsen TE, Andersen JT, and Högberg B (2019). Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies. Nat. Nanotechnol 14, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veneziano R, Moyer TJ, Stone MB, Wamhoff E-C, Read BJ, Mukherjee S, Shepherd TR, Das J, Schief WR, Irvine DJ, and Bathe M (2020). Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol 15, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kato Y, Abbott RK, Freeman BL, Haupt S, Groschel B, Silva M, Menis S, Irvine DJ, Schief WR, and Crotty S (2020). Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 53, 548–563.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marcandalli J, Fiala B, Ols S, Perotti M, de van der Schueren W, Snijder J, Hodge E, Benhaim M, Ravichandran R, Carter L, et al. (2019). Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell 176, 1420–1431.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benton DJ, Gamblin SJ, Rosenthal PB, and Skehel JJ (2020). Structural transitions in influenza haemagglutinin at membrane fusion pH. Nature 583, 150–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das DK, Govindan R, Nikic-Spiegel I, Krammer F, Lemke EA, and Munro JB (2018). Direct Visualization of the Conformational Dynamics of Single Influenza Hemagglutinin Trimers. Cell 174, 926–937.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang P-S, Oberdorfer G, Xu C, Pei XY, Nannenga BL, Rogers JM, DiMaio F, Gonen T, Luisi B, and Baker D (2014). High thermo-dynamic stability of parametrically designed helical bundles. Science 346, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lupas AN, Bassler J, and Dunin-Horkawicz S (2017). The Structure and Topology of α-Helical Coiled Coils. Subcell. Biochem 82, 95–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woolfson DN (2005). The design of coiled-coil structures and assemblies. Adv. Protein Chem 70, 79–112. [DOI] [PubMed] [Google Scholar]
- 68.Harbury PB, Kim PS, and Alber T (1994). Crystal structure of an isoleucine-zipper trimer. Nature 371, 80–83. [DOI] [PubMed] [Google Scholar]
- 69.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RMB, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. (2015). A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306. [DOI] [PubMed] [Google Scholar]
- 70.Ueda G, Antanasijevic A, Fallas JA, Sheffler W, Copps J, Ellis D, Hutchinson GB, Moyer A, Yasmeen A, Tsybovsky Y, et al. (2020). Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife 9, e57659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, Alam N, Al-ford RF, Aprahamian M, Baker D, Barlow KA, Barth P, et al. (2020). Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods 17, 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boyoglu-Barnum S, Ellis D, Gillespie RA, Hutchinson GB, Park Y-J, Moin SM, Acton OJ, Ravichandran R, Murphy M, Pettie D, et al. (2021). Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antanasijevic A, Ueda G, Brouwer PJM, Copps J, Huang D, Allen JD, Cottrell CA, Yasmeen A, Sewall LM, Bontjer I, et al. (2020). Structural and functional evaluation of de novo-designed, two-component nanoparticle carriers for HIV Env trimer immunogens. PLoS Pathog. 16, e1008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sliepen K, Radic L, Capella-Pujol J, Watanabe Y, Zon I, Chumbe A, Lee W-H, de Gast M, Koopsen J, Koekkoek S, et al. (2022). Induction of cross-neutralizing antibodies by a permuted hepatitis C virus glycoprotein nanoparticle vaccine candidate. Nat. Commun 13, 7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malhi H, Homad LJ, Wan Y-H, Poudel B, Fiala B, Borst AJ, Wang JY, Walkey C, Price J, Wall A, et al. (2022). Immunization with a self-assembling nanoparticle vaccine displaying EBV gH/gL protects humanized mice against lethal viral challenge. Cell Rep. Med 3, 100658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walls AC, Fiala B, Schäfer A, Wrenn S, Pham MN, Murphy M, Tse LV, Shehata L, O’Connor MA, Chen C, et al. (2020). Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brouwer PJM, Antanasijevic A, Ronk AJ, Muller-Kräüter H, Watanabe Y, Claireaux M, Perrett HR, Bijl TPL, Grobben M, Umotoy JC, et al. (2022). Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection. Cell Host Microbe 30, 1759–1772.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laver WG, and Valentine RC (1969). Morphology of the isolated hemagglutinin and neuraminidase subunits of influenza virus. Virology 38, 105–119. [DOI] [PubMed] [Google Scholar]
- 79.McCraw DM, Gallagher JR, and Harris AK (2016). Characterization of Influenza Vaccine Hemagglutinin Complexes by Cryo-Electron Microscopy and Image Analyses Reveals Structural Polymorphisms. Clin. Vaccine Immunol 23, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dosey A, Ellis D, Boyoglu-Barnum S, Syeda H, Saunders M, Watson M, Kraft JC, Pham MN, Guttman M, Lee KK, et al. Combinatorial Immune Refocusing within the Influenza Hemagglutinin Head Elicits Cross-Neutralizing Responses. [DOI] [PMC free article] [PubMed]
- 81.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tokatlian T, Read BJ, Jones CA, Kulp DW, Menis S, Chang JYH, Steichen JM, Kumari S, Allen JD, Dane EL, et al. (2019). Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 363, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin JT, Cottrell CA, Antanasijevic A, Carnathan DG, Cossette BJ, Enemuo CA, Gebru EH, Choe Y, Viviano F, Fischinger S, et al. (2020). Targeting HIV Env immunogens to B cell follicles in nonhuman primates through immune complex or protein nanoparticle formulations. NPJ Vaccines 5, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Read BJ, Won L, Kraft JC, Sappington I, Aung A, Wu S, Bals J, Chen C, Lee KK, Lingwood D, et al. (2022). Mannose-binding lectin and complement mediate follicular localization and enhanced immunogenicity of diverse protein nanoparticle immunogens. Cell Rep. 38, 110217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cohen AA, Gnanapragasam PNP, Lee YE, Hoffman PR, Ou S, Kakutani LM, Keeffe JR, Wu H-J, Howarth M, West AP, et al. (2021). Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dalvie NC, Rodriguez-Aponte SA, Hartwell BL, Tostanoski LH, Biedermann AM, Crowell LE, Kaur K, Kumru OS, Carter L, Yu J, et al. (2021). Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proc. Natl. Acad. Sci. USA 118, e2106845118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song JY, Choi WS, Heo JY, Lee JS, Jung DS, Kim S-W, Park K-H, Eom JS, Jeong SJ, Lee J, et al. (2022). Safety and immunogenicity of a SARS-CoV-2 recombinant protein nanoparticle vaccine (GBP510) adjuvanted with AS03: A randomised, placebo-controlled, observer-blinded phase 1/2 trial. EClinicalMedicine 51, 101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song JY, Choi WS, Heo JY, Kim EJ, Lee JS, Jung DS, Kim S-W, Park K-H, Eom JS, Jeong SJ, et al. (2023). Immunogenicity and safety of SARS-CoV-2 recombinant protein nanoparticle vaccine GBP510 adjuvanted with AS03: randomised, active-controlled, observer-blinded, phase 3 trial. Preprint at medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanekiyo M, Bu W, Joyce MG, Meng G, Whittle JRR, Baxa U, Yamamoto T, Narpala S, Todd J-P, Rao SS, et al. (2015). Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 162, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perotti M, Marcandalli J, Demurtas D, Sallusto F, and Perez L (2020). Rationally designed Human Cytomegalovirus gB nanoparticle vaccine with improved immunogenicity. PLoS Pathog. 16, e1009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellis D, Brunette N, Crawford KHD, Walls AC, Pham MN, Chen C, Herpoldt K-L, Fiala B, Murphy M, Pettie D, et al. (2021). Stabilization of the SARS-CoV-2 Spike Receptor-Binding Domain Using Deep Mutational Scanning and Structure-Based Design. Front. Immunol 12, 710263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McLeod B, Mabrouk MT, Miura K, Ravichandran R, Kephart S, Hailemariam S, Pham TP, Semesi A, Kucharska I, Kundu P, et al. (2022). Vaccination with a structure-based stabilized version of malarial antigen Pfs48/45 elicits ultra-potent transmission-blocking antibody responses. Immunity 55, 1680–1692.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jardine J, Julien J-P, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, MacPherson S, Jones M, et al. (2013). Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duan H, Chen X, Boyington JC, Cheng C, Zhang Y, Jafari AJ, Stephens T, Tsybovsky Y, Kalyuzhniy O, Zhao P, et al. (2018). Glycan Masking Focuses Immune Responses to the HIV-1 CD4-Binding Site and Enhances Elicitation of VRC01- Class Precursor Antibodies. Immunity 49, 301–311.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leggat DJ, Cohen KW, Willis JR, Fulp WJ, deCamp AC, Kalyuzhniy O, Cottrell CA, Menis S, Finak G, Ballweber-Fleming L, et al. (2022). Vaccination induces HIV broadly neutralizing antibody precursors in humans. Science 378, eadd6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen AA, van Doremalen N, Greaney AJ, Andersen H, Sharma A, Starr TN, Keeffe JR, Fan C, Schulz JE, Gnanapragasam PNP, et al. (2022). Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science 377, eabq0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walls AC, Miranda MC, Schäfer A, Pham MN, Greaney A, Arunachalam PS, Navarro M-J, Tortorici MA, Rogers K, O’Connor MA, et al. (2021). Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell 184, 5432–5447.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yassine HM, Boyington JC, McTamney PM, Wei C-J, Kanekiyo M, Kong W-P, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. (2015). Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med 21, 1065–1070. [DOI] [PubMed] [Google Scholar]
- 99.Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, et al. (2019). Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsieh C-L, Werner AP, Leist SR, Stevens LJ, Falconer E, Goldsmith JA, Chou C-W, Abiona OM, West A, Westendorf K, et al. (2021). Stabilized coronavirus spike stem elicits a broadly protective antibody. Cell Rep. 37, 109929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bowen JE, Park Y-J, Stewart C, Brown JT, Sharkey WK, Walls AC, Joshi A, Sprouse KR, McCallum M, Tortorici MA, et al. (2022). SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines. Sci. Immunol 7, eadf1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ellis D, Lederhofer J, Acton OJ, Tsybovsky Y, Kephart S, Yap C, Gillespie RA, Creanga A, Olshefsky A, Stephens T, et al. (2022). Structure-based design of stabilized recombinant influenza neuraminidase tetramers. Nat. Commun 13, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Milder FJ, Jongeneelen M, Ritschel T, Bouchier P, Bisschop IJM, de Man M, Veldman D, Le L, Kaufmann B, Bakkers MJG, et al. (2022). Universal stabilization of the influenza hemagglutinin by structure-based redesign of the pH switch regions. Proc. Natl. Acad. Sci. USA 119, e2115379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Joyce MG, Zhang B, Ou L, Chen M, Chuang G-Y, Druz A, Kong W-P, Lai Y-T, Rundlet EJ, Tsybovsky Y, et al. (2016). Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat. Struct. Mol. Biol 23, 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Che Y, Gribenko AV, Song X, Handke LD, Efferen KS, Tompkins K, Kodali S, Nunez L, Prasad AK, Phelan LM, et al. (2023). Rational Design of a Highly Immunogenic Prefusion-Stabilized F Glycoprotein Antigen for a Respiratory Syncytial Virus Vaccine. Sci. Transl. Med 15, eade6422. [DOI] [PubMed] [Google Scholar]
- 106.Stewart-Jones GBE, Gorman J, Ou L, Zhang B, Joyce MG, Yang L, Cheng C, Chuang G-Y, Foulds KE, Kong W-P, et al. (2021). Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses. Proc. Natl. Acad. Sci. USA 118, e2106196118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hsieh C-L, Rush SA, Palomo C, Chou C-W, Pickens W, Más V, and McLellan JS (2022). Structure-based design of prefusion-stabilized human metapneumovirus fusion proteins. Nat. Commun 13, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chuang G-Y, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, et al. (2017). Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity. J. Virol 91, e02268–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J-P, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. (2015). Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell 163, 1702–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rutten L, Lai Y-T, Blokland S, Truan D, Bisschop IJM, Strokappe NM, Koornneef A, van Manen D, Chuang G-Y, Farney SK, et al. (2018). A Universal Approach to Optimize the Folding and Stability of Prefusion-Closed HIV-1 Envelope Trimers. Cell Rep 23, 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu D, Li C, Utz A, Weidenbacher PAB, Tang S, Sanyal M, Pulendran B, and Kim PS (2022). Designing epitope-focused vaccines via antigen reorientation. Preprint at bioRxiv. [Google Scholar]
- 112.Frey SJ, Carreño JM, Bielak D, Arsiwala A, Altomare CG, Varner C, Rosen-Cheriyan T, Bajic G, Krammer F, and Kane RS (2023). Nanovaccines Displaying the Influenza Virus Hemagglutinin in an Inverted Orientation Elicit an Enhanced Stalk-Directed Antibody Response. Adv. Healthcare Mater 12, e2202729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Y, Huynh DT, and Yeates TO (2019). A 3.8Å resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nat. Commun 10, 1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vulovic I, Yao Q, Park Y-J, Courbet A, Norris A, Busch F, Sahasrabuddhe A, Merten H, Sahtoe DD, Ueda G, et al. (2021). Generation of ordered protein assemblies using rigid three-body fusion. Proc. Natl. Acad. Sci. USA 118, e2015037118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oktay E, Alem F, Hernandez K, Girgis M, Green C, Mathur D, Medintz IL, Narayanan A, and Veneziano R (2023). DNA origami presenting the receptor binding domain of SARS-CoV-2 elicit robust protective immune response. Commun. Biol 6, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wamhoff E-C, Ronsard L, Feldman J, Knappe GA, Hauser BM, Romanov A, Lam E, Denis KS, Boucau J, Barczak AK, et al. (2023). Enhancing antibody responses by multivalent antigen display on thymus-independent DNA origami scaffolds. Preprint at bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 46, 1073–1088.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. (2019). Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 177, 1153–1171.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aung A, Cui A, Maiorino L, Amini AP, Gregory JR, Bukenya M, Zhang Y, Lee H, Cottrell CA, Morgan DM, et al. (2023). Low protease activity in B cell follicles promotes retention of intact antigens after immunization. Science 379, eabn8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dauparas J, Anishchenko I, Bennett N, Bai H, Ragotte RJ, Milles LF, Wicky BIM, Courbet A, de Haas RJ, Bethel N, et al. (2022). Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lutz ID, Wang S, Norn C, Courbet A, Borst AJ, Zhao YT, Dosey A, Cao L, Xu J, Leaf EM, et al. (2023). Top-down design of protein architectures with reinforcement learning. Science 380, 266–273. [DOI] [PubMed] [Google Scholar]
- 123.Watson JL, Juergens D, Bennett NR, Trippe BL, Yim J, Eisenach HE, Ahern W, Borst AJ, Ragotte RJ, Milles LF, et al. (2023). De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weaver GC, Villar RF, Kanekiyo M, Nabel GJ, Mascola JR, and Lingwood D (2016). In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat. Protoc 11, 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kratochvil S, Shen C-H, Lin Y-C, Xu K, Nair U, Da Silva Pereira L, Tripathi P, Arnold J, Chuang G-Y, Melzi E, et al. (2021). Vaccination in a humanized mouse model elicits highly protective PfCSP-targeting anti-malarial antibodies. Immunity 54, 2859–2876.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Melzi E, Willis JR, Ma KM, Lin Y-C, Kratochvil S, Berndsen ZT, Landais EA, Kalyuzhniy O, Nair U, Warner J, et al. (2022). Membrane-bound mRNA immunogens lower the threshold to activate HIV Env V2 apex-directed broadly neutralizing B cell precursors in humanized mice. Immunity 55, 2168–2186.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luo S, Jing C, Ye AY, Kratochvil S, Cottrell CA, Koo J-H, Chap-delaine Williams A, Francisco LV, Batra H, Lamperti E, et al. (2023). Humanized V(D)J-rearranging and TdT-expressing mouse vaccine models with physiological HIV-1 broadly neutralizing antibody precursors. Proc. Natl. Acad. Sci. USA 120, e2217883120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang X, Ray R, Kratochvil S, Melzi E, Lin Y-C, Giguere S, Xu L, Warner J, Cheon D, Liguori A, et al. (2021). Multiplexed CRISPR/CAS9-mediated engineering of pre-clinical mouse models bearing native human B cell receptors. EMBO J. 40, e105926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang P-S, Ban Y-EA, Richter F, Andre I, Vernon R, Schief WR, and Baker D (2011). RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One 6, e24109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. (2014). Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol 88, 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Barouch DH, Yang Z-Y, Kong W-P, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, et al. (2005). A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol 79, 8828–8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60. [DOI] [PubMed] [Google Scholar]
- 133.Tegunov D, and Cramer P (2019). Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- 135.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lin-dahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zivanov J, Nakane T, and Scheres SHW (2019). A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ilca SL, Kotecha A, Sun X, Poranen MM, Stuart DI, and Huiskonen JT (2015). Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun 6, 8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abrishami V, Ilca SL, Gomez-Blanco J, Rissanen I, de la Rosa-Trevín JM, Reddy VS, Carazo J-M, and Huiskonen JT (2021). Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Prog. Biophys. Mol. Biol 160, 43–52. [DOI] [PubMed] [Google Scholar]
- 139.Chen S, McMullan G, Faruqi AR, Murshudov GN, Short JM, Scheres SHW, and Henderson R (2013). High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- 141.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 142.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Frenz B, Rämisch S, Borst AJ, Walls AC, Adolf-Bryfogle J, Schief WR, Veesler D, and DiMaio F (2019). Automatically Fixing Errors in Glycoprotein Structures with Rosetta. Structure 27, 134–139.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang RY-R, Song Y, Barad BA, Cheng Y, Fraser JS, and DiMaio F (2016). Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, e17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.World Health Organization, and WHO Global Influenza Surveillance Network (2011). Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza (World Health Organization; ). [Google Scholar]
- 147.Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, Hatch L, Stephens T, Tsybovsky Y, Crank MC, et al. (2021). A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nat. Commun 12, 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural data generated from this study were deposited in public data repositories and the accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| C05 | Produced in house (Ekiert et al.)22 | N/A |
| FluA-20 | Produced in house (Bangaru et al.)29 | N/A |
| Ab6649 | Produced in house (Raymond et al.)35 | N/A |
| Goat anti-human IgG, HRP | SouthernBiotech | Cat# 2040–05; RRID:AB_2795644 |
| Goat anti-mouse IgG, HRP | SouthernBiotech | Cat# 1080–05; RRID:AB_2734756 |
| Bacterial and virus strains | ||
| A/New Caledonia/20/1999 R3ΔPB1 virus | Produced in house | N/A |
| Influenza reporter viruses | Produced in house (Creanga et al.)147 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| AddaVax adjuvant | Cat# vac-adx-10 | |
| TMB | SeraCare | Cat# 5120–0083 |
| Polyethylenimine (PEI) HCl MAX, Linear, Mw 40,000 | Polysciences | Cat# 24765–1 |
| PNGase F | NEB | Cat# P0705 |
| RDE II | VWR | Cat# 10753–482 |
| Deposited data | ||
| Trihead-1heptad-I53_dn5 | EMDataBank | EMD: 42481 |
| Localized reconstruction of Trihead-1heptad-I53_dn5 | PDB, EMDataBank | PDB ID: 8UR5, EMD: 42482 |
| Trihead-2heptad-I53_dn5 | EMDataBank | EMD: 42483 |
| Trihead-6heptad-I53_dn5 | EMDataBank | EMD: 42485 |
| Localized reconstruction of Trihead-6heptad-I53_dn5 | PDB, EMDataBank | PDB ID: 8UR7, EMD:42486 |
| Experimental models: Cell lines | ||
| Expi293F | ThermoFisher | Cat# A14527 |
| MDCK-SIAT-PB1 | Produced in house (Creanga et al.)147 | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c mice | Jackson Laboratory | Cat# 000651 |
| Recombinant DNA | ||
| See Table S1 for amino acid sequences | ||
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Leginon | (Suloway et al.)132 | N/A |
| CryoSparc | (Punjanietal.)134 | https://cryosparc.com/ |
| UCSF Chimera | (Pettersen et al.)141 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX | (Goddard et al.)145 | https://www.rbvi.ucsf.edu/chimerax/ |
| PyMol | Schrodinger, LLC | https://pymol.org/2/ |
| Unicorn 7.0 | GE Healthcare | http://www.gelifesciences.com/ |
| Relion | (Zivanov et al.)135 | N/A |
| Other | ||
| 300 mesh grids | Ted Pella | Cat# 01843-F |
| UltrAufoil 2/2, 200 mesh grids | Electron Microscopy Sciences | |
| Filter paper | Cytiva | Cat# 1004047 |
| Uranyl formate | SPI Chem | Cat# 02545-AA |
| Superdex 200 Increase SEC column | Cytiva | Cat# 28-9909-44 |
| Superose 6 Increase SEC column | Cytiva | Cat# 29091596 |
| Excel resin | Cytiva | Cat# 17371203 |
| Isoflurane | USP Patterson | Cat# 07-893-1389 |
| EndoSafe LAL Test Cartridges | Charles River Labs | Cat# PTS20005F |
| Lemo21(DE3) | New England BioLabs | Cat# C2528J |
| MAX Efficiency™ DH5α Competent Cells | Sigma-Aldrich | Cat# I6758 |
| Isopropyl-B-D-thiogalactoside (IPTG) | Thermo Fisher | Cat# 18258012 |
| Kanamycin Sulfate | Sigma-Aldrich | Cat# K1876 |
| HisTrap HP | Cytiva | Cat#17-5247-01 |
| Octet Protein A Biosensors | Sartorius | Cat# 18-5010 |
| Turkey Red Blood Cells | Lampire | Cat# # 7209403 |
| Plasmid Plus Maxi Kit | Qiagen | Cat# 12963 |
| Trypsin, TPCK Treated | Thermo Fisher | Cat# 20233 |


