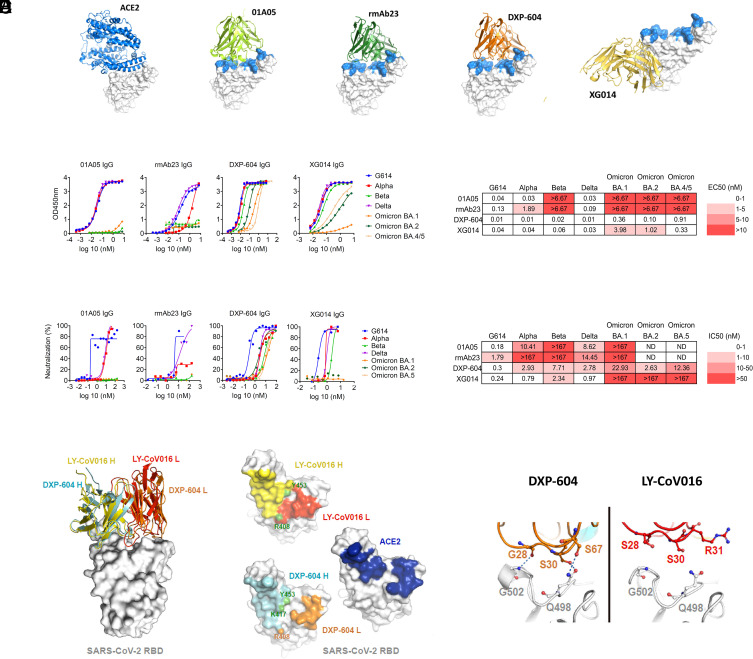
Fig. 2.
Characterization of neutralizing antibodies 01A05, rmAb23, DXP-604, and XG014. (A) In silico binding of ACE2 and IgG antibodies to RBD. The ACE2 receptor binding motif is indicated (blue). (B and C) Binding to RBD (B) and neutralization (C) of G614 and VOCs by IgG antibodies. The EC50 and IC50 and fold-change differences between IgG and IgA antibody forms are indicated. (D) Overlaid crystal structures of LY-CoV016 Fab (PDB ID: 7C01) and DXP-604 Fab 473 (PDB ID: 7CH4) in complex with SARS-CoV-2 RBD (Left picture) and the footprints of LY-CoV016, DXP-604, and ACE2 (PDB ID: 6M0J) on SARS-CoV-2 RBD. Atoms of the RBD within 5.0 Å of the antibodies or ACE2 are colored yellow (LY-CoV016 H), red (LY-CoV016 L), cyan (DXP-604 H), orange (DXP-604 L), or blue (ACE2) (Right picture). (E) Hydrogen bonds were formed between S30/S67 in the light chain of DXP-604 and RBD Q498, which is a key ACE2-binding site, and between the main chain groups of G28 and RBD G502. See also SI Appendix, Figs. S2–S4.