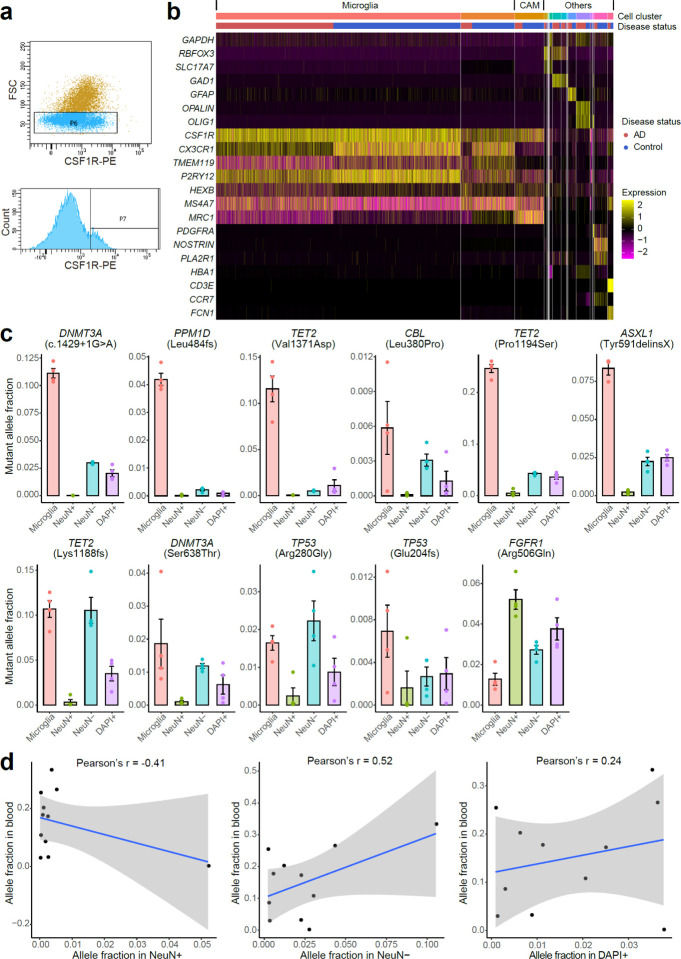
Extended Data Fig. 4. Microglial purity and mutant allele fraction of FANS-sorted nuclei population.
a, Selectively isolated microglia from frozen brain tissues using FANS with an antibody targeting epitopes of CSF1R, a gene highly expressed in microglia. b, Marker gene expression profile for 10X single-nucleus RNA-seq of CSF1R+ sorted nuclei. Each column represents a single nucleus, clustered by PCA based on their expression similarity. About 75–77% of the sorted nuclei are microglia with high expression of CX3CR1, TMEM119, and P2RY12, whereas another 4–9% are CNS-associated macrophages (CAMs). Markers for blood cell types (HBA1: red blood cell; CD3E: T cell; CCR7: B cell; FCN1: monocyte) confirm the minimal presence of blood cells in sorted nuclei. CNS, central nervous system. AD microglia showed generally reduced expression of CX3CR1 and P2RY12, consistent with previous findings in AD3. c, Mutant allele fractions across different sorted nuclei populations for all the 11 profiled AD somatic mutations. Four mutations are shown in Fig. 4c as examples. In all but the FGFR1 mutation, we observed significantly higher allele fractions in microglia than in neurons (NeuN+). Each population of nuclei was sorted four times from each AD brain sample to serve as replicates. Error bar, SE. d, The correlation of mutant allele fractions between blood and three nuclei populations (NeuN+, NeuN−, and DAPI+) sorted from matched brain samples.