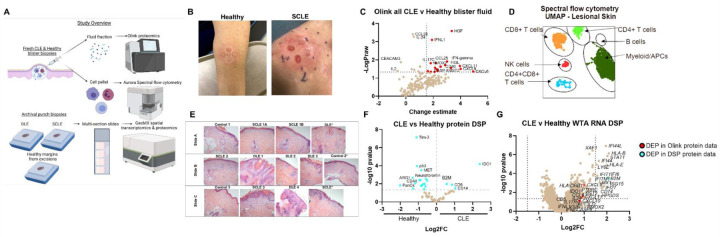
Fig. 1. Integration of high-resolution immune techniques with spatial approaches for studying cutaneous lupus erythematosus.
(A) Study overview demonstrating workflow. We performed blister biopsies on cutaneous lupus erythematosus patients and healthy donors and analyzed the interstitial skin fluid using Olink proteomics and the cell pellets using Aurora spectral flow cytometry. We also employed biobanked FFPE skin biopsies from DLE, SCLE and healthy margins for NanoString digital spatial profiling for both protein, using 6 modules, and RNA, using the whole transcriptome assay (WTA). Created with Biorender.com. (B) Sample photographs of blister biopsies (n=3 healthy and 4 CLE patients). (C) Volcano plot of Olink data from the inflammation and neuroexploratory panels of all CLE blisters versus healthy controls (n=7 CLE lesional, 7 CLE nonlesional, and 3 healthy blisters). (D) Sample UMAP of immune infiltrates in CLE lesional skin. (E) H&E images of the slide set used for spatial transcriptomics and proteomics. (n=3 healthy margin controls, 5 SCLE biopsies from 4 patients, and 4 DLE biopsies from 4 patients). (F) Volcano plot of DSP protein data for all CLE regions of interest (ROIs) versus healthy margin ROIs. (G) Volcano plot of DSP RNA WTA data for all CLE ROIs versus healthy margin ROIs. Red dots are also differentially expressed proteins (DEPs) in the Olink protein dataset, and cyan dots are also DEPs in the DSP protein dataset. (n=41 ROIs assessed from 4 DLE, 3 SCLE, and 2 healthy margin control biopsies for WTA DSP; n=95 ROIs assessed from 4 DLE, 4 SCLE from 3 donors and 3 healthy margin controls for protein DSP. Discrepancy due to tissue not being fully seated in imaging area for WTA run, indicated by *.)