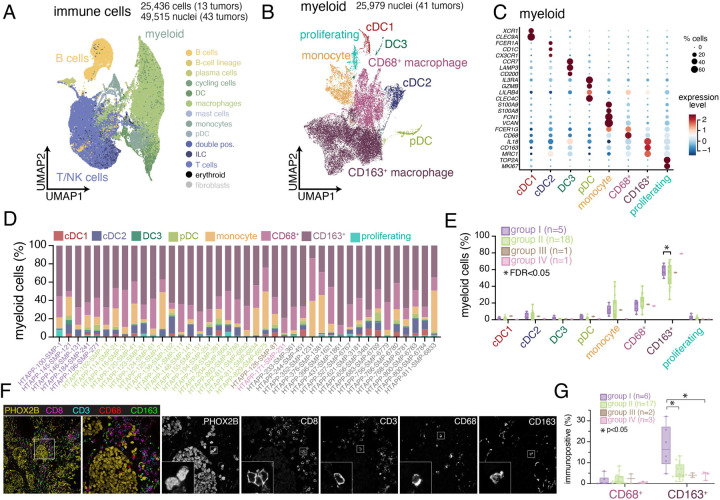
Figure 5. Immune cell heterogeneity and spatial compartmentalization of neuroblastoma.
(A) UMAP plot of integrated immune cells/nuclei from scRNA-seq (n=13 tumors; 25,436 cells) and snRNA-seq (n=43 tumors; 49,515 nuclei). Cells/nuclei are colored based on CellTypist automated annotation75.
(B) UMAP plot of 25,979 myeloid nuclei from the HTAPP neuroblastoma dataset (B). Nuclei are categorized based on the expression of cell markers.
(C) Dot plot showing expression of cell markers for each myeloid cluster. Expression is colored based on scaled normalized value (z-score) and the size of each dot represents the percentage of cells within each cluster that had detectable expression.
(D) Bar plots showing cell type proportions of myeloid subtypes for each sample within the snRNA-seq cohort (n=41).
(E) Composition analysis of the myeloid compartment across snRNA-seq datasets (n=41), separated by methylation grouping. Data are presented as median ± interquartile range. Statistically credible differences, as measured using Bayesian component analysis with false discover rate < 0.05, are represented with an asterisk.
(F) Example image of multiplexed immunofluorescence for sample HTAPP-102-SMP-11, stained with PHOX2B, CD8, CD3, CD68, and CD163.
(G) Quantitation of CD68+ and CD163+ cells from multiplexed immunofluorescence (n=28). Data is divided by methylation grouping. Data is presented as median ± interquartile range. Differences across groups that meet statistical significance are marked with an asterisk (p values; Wilcoxon rank sum test).