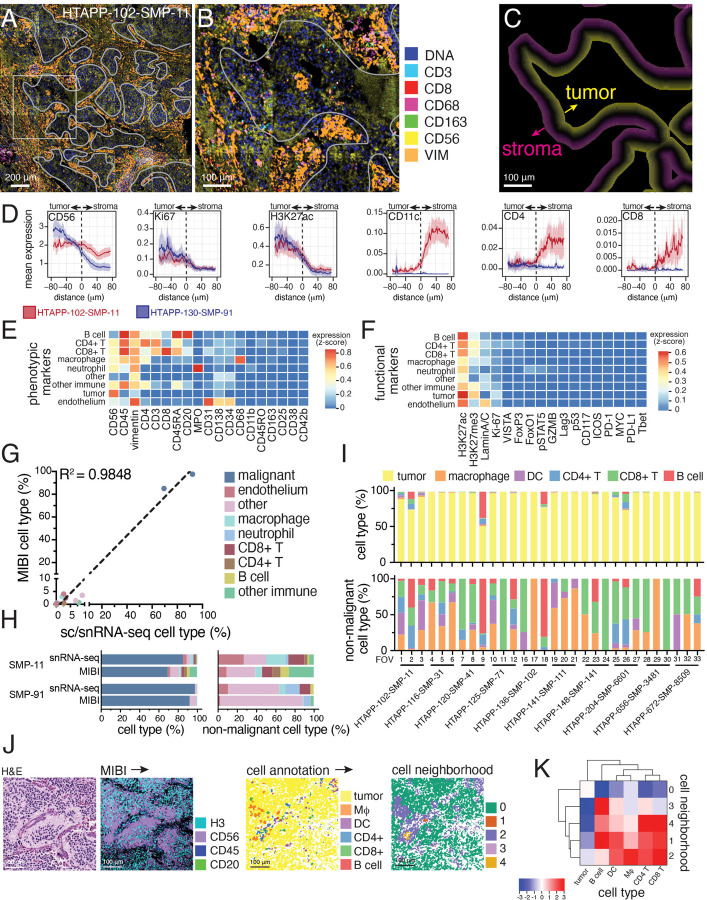
Figure 6. Multiplexed ion-beam imaging (MIBI) identifies compartmentalization of tumor and immune cells.
(A) Multiplexed ion beam imaging (MIBI) of multiplexed spatial proteomic data. Two large, tiled arrays (5x5 captured areas) from samples HTAPP-102-SMP-11 and HTAPP-130-SMP-91 were obtained. As an example, the stitched array from HTAPP-102-SMP-11 is shown in panel A; 7 markers are shown (double-stranded DNA [dsDNA], CD3, CD8, CD68, CD163, CD56 and Vimentin [VIM]). A white overlay shows the tumor-stroma interface, which was used for patched level analysis of neuroblastoma (PLANB).
(B) A high magnification of the boxed area within (A).
(C) Patched level analysis of neuroblastoma (PLANB) determines the relative locality of cell types or markers with reference to the tumor-stroma boundary using a stepwise pixel-by-pixel algorithm.
(D) PLANB analysis of HTAPP-102-SMP-11 and HTAPP-130-SMP-91 showing the distribution of tumor markers (CD56, Ki67, H3K27ac), myeloid cells (CD11c), or T cells (CD4, CD8). The x-axis reflects distance relative to tumor-stroma interface, with negative values being within the tumor nest and positive values being outside the tumor nest. The y-axis reflects relative expression of markers (arbitrary units).
(E-F) Heatmap showing expression of cell type phenotypic markers (E) and functional markers (F) for each cell type in the large, tiled arrays.
(G-H) Cell type proportions from the two large, tiled arrays comparing the prevalence of cell types in the MIBI data compared to snRNA-seq data. Panel (G) shows a scatterplot comparing proportions of each cell type identified in sc/snRNA-seq data (x-axis) to the proportion detected in the tiled MIBI arrays for HTAPP-102-SMP-11 and HTAPP-130-SMP-91. Panel (H) shows bar plots comparing cell type composition in snRNA-seq and MIBI datasets both including (left) and excluding (right) malignant cells/nuclei.
(I) Bar plot of cell types in the single-tile MIBI datasets (10 samples, 30 FOVs). Following capture of MIBI data, segmentation and marker expression was used to annotate cell types. Bar plots including and excluding malignant cells are shown on the top and bottom, respectively.
(J) Neighborhood analysis of MIBI data from the 30 FOVs from (H). As an example, we show HTAPP-102-SMP-11 field-of-view (FOV) #1. On the left side, we show an H&E from an adjacent section. Following MIBI capture, images were segmented, and cell types were automatically called. These cell maps were then analyzed using neighborhood analysis to identify those cell types that co-localize with each other.
(K) Cluster-map of cell type composition within each neighborhood from the combined MIBI dataset (n=30 FOVs). Dendrograms represent hierarchical clustering of cell types (rows) and neighborhoods (columns). Colors represent scaled likelihood of adjacency between cell types.