Figure 3. HKU1 RBD binding inhibits human TMPRSS2 activity.
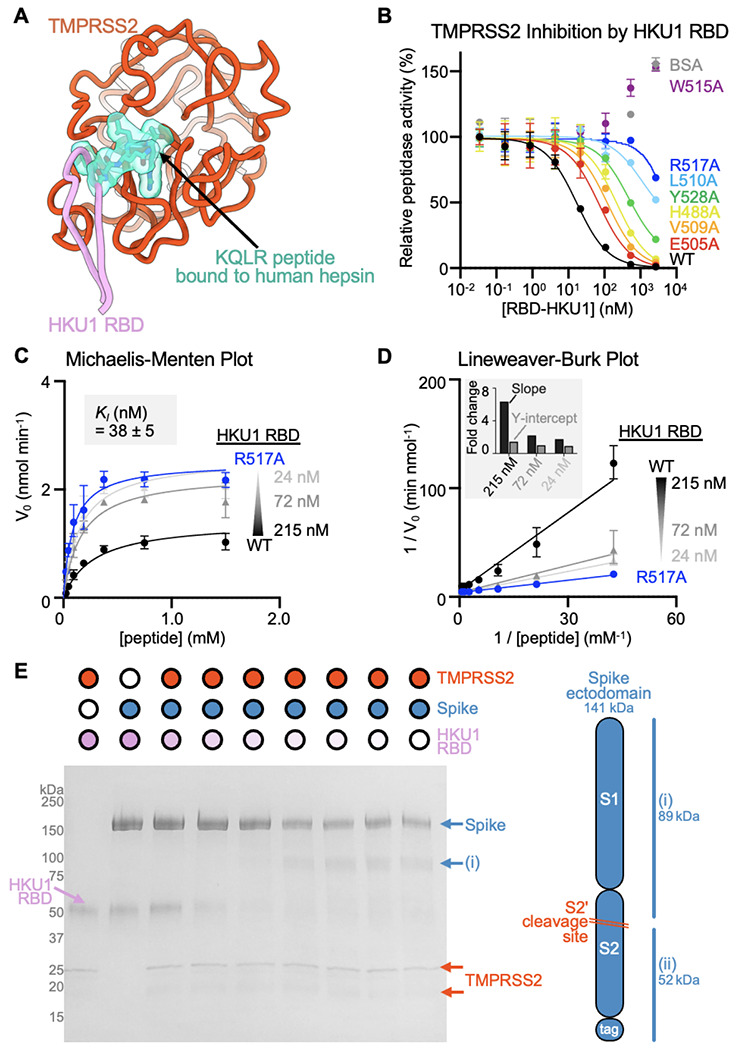
(A) Superimposition of the KQLR chloromethylketone-bound hepsin structure (PDB ID 1Z8G46) to the HKU1-bound TMPRSS2 structure showing that the ligand would be precluded sterically from binding in the active site upon attachment of the HKU1 RBD (assuming an identical binding mode of the ligand). Only the TMPRSS2 peptidase domain is shown and hepsin is omitted for clarity. (B) Assessment of inhibition of human TMPRSS2 activity by the wildtype (WT) isolate N1 and interface mutant HKU1 RBDs using the fluorescent Boc-QAR-AMC peptide substrate in the presence of 3.4 nM of the TMPRSS2 ectodomain harboring the DS, SUMO, and N249 glycan. Data are shown as the geometric mean and standard deviation of 2-3 technical replicates. (C-D) Michaelis–Menten plot (C) and Lineweaver-Burke plot (D) of initial reaction velocities in function of the concentration of fluorescent Boc-QAR-AMC peptide substrate, at 22 °C, in the presence of 5 nM TMPRSS2 and the wildtype (WT) isolate N1 or R517A interface mutant HKU1 RBDs. The inhibitor constant KI was determined through fitting competitive inhibition using GraphPad Prism. Data are shown as the geometric mean and standard deviation of three technical replicates. The inset in panel D shows the relative change in slope (KM/Vmax) and y axis intercept (1/Vmax) for the different concentrations of wildtype HKU1 RBD relative to the R517A RBD. (E) Reducing SDS-PAGE analysis of proteolytic processing at 37°C of 5 μg of SARS-CoV-2 S 2P (S2P) by 0.5 μM TMPRSS2 preincubated with various concentrations of the HKU1 isolate N1 RBD.
