Extended Data Fig. 1.
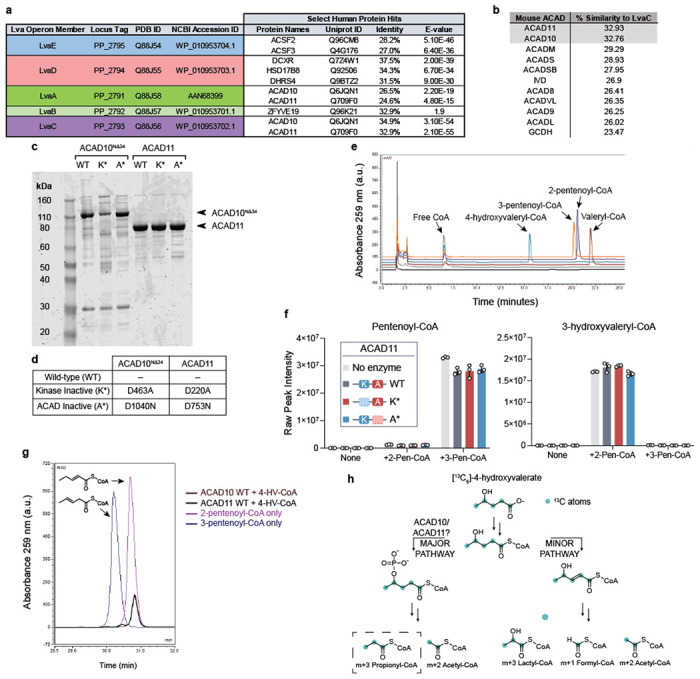
(a) Sequence identity of lva operon enzymes to human proteins as reported by UniProtKB database. (b) Sequence similarity of LvaC relative to mouse ACAD family according to pairwise alignments. (c) SDS-PAGE gel of recombinant mouse ACAD10NΔ34 and ACAD11 protein preparations from P. pastoris or RIPL E. coli orthogonal expression systems, respectively. WT = wild type; K* = Kinase inactive mutant; A* = ACAD inactive mutant. (d) Table describing the site-specific mutations to inactivate recombinant ACAD10NΔ34 and ACAD11 APH and ACAD catalytic domains. (e) HPLC chromatogram of acyl-CoA standards. (f) Assay testing the hydration activity of recombinant ACAD11. 2-pentenoyl-CoA (2-Pen-CoA) and 3-pentenoyl-CoA (3-Pen-CoA) were tested as substrates. Raw signal intensities of peaks corresponding to substrates and 3-hydroxyvaleryl-CoA product are depicted. Data represents mean −/+ SD (n = 3 technical replicates). (g) HPLC trace of pentenoyl-CoA isomers formed from 500 μM 4-HV-CoA by recombinant wild-type ACAD10NΔ34 and ACAD11 in vitro (brown and black traces, respectively) in comparison to LvaE-generated pentenoyl-CoA standards (dark purple and blue traces, respectively). (h) Expected labeling pattern of acyl-CoA catabolites derived from the catabolism of [13C5]-4-hydroxyvalerate (4-HV) by the “major” or “minor” pathways. M+3 propionyl-CoA labeling reflects catabolism through the 4-phosphovaleryl-CoA intermediate, presumably catalyzed by ACAD10/11.
