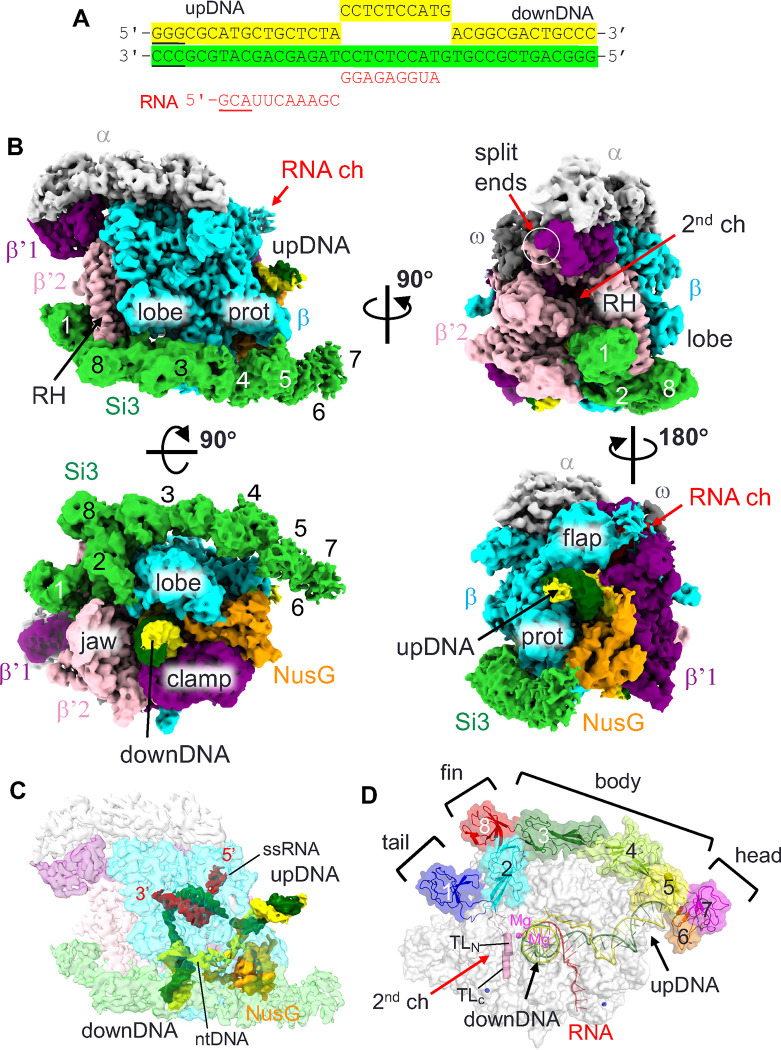
Fig. 2. Cryo-EM image of the cyRNAP elongation complex with NusG.
(A) The sequence of the DNA/RNA scaffold used for the EC-NusG assembly (template DNA, green; nontemplate DNA, yellow; RNA, red). DNA and RNA regions lacking cryo-EM density are underlined. (B) Orthogonal views of the cryo-EM density map. Subunits and domains of cyRNAP, DNA, RNA and NusG are colored and labeled (RH, rim helix; prot, protrusion; downDNA, downstream DNA; upDNA, upstream DNA). The split ends of the β’1 and β’2 subunits are indicated by white circles. The SBHMs in Si3 are labeled 1 to 8. (C) Cryo-EM density of DNA, RNA and NusG are shown with a transparent RNAP density map (ntDNA, nontemplate DNA; ssRNA, single-stranded RNA). The 5’ and 3’ ends of the RNA are indicated. The cryo-EM density map is colored according to B. (D) Efficient storage of an elongated and large Si3 molecule on the surface of cyRNAP. The structure of EC-NusG is shown as a transparent surface, and the Si3, DNA/RNA and trigger loop (TLN, TLC) regions are shown as cartoon models. SBHMs are labeled 1 to 8, and subdomains (tail, fin, body and head) are indicated. The active site of RNAP is designated by catalytic Mg2+ (magenta sphere).