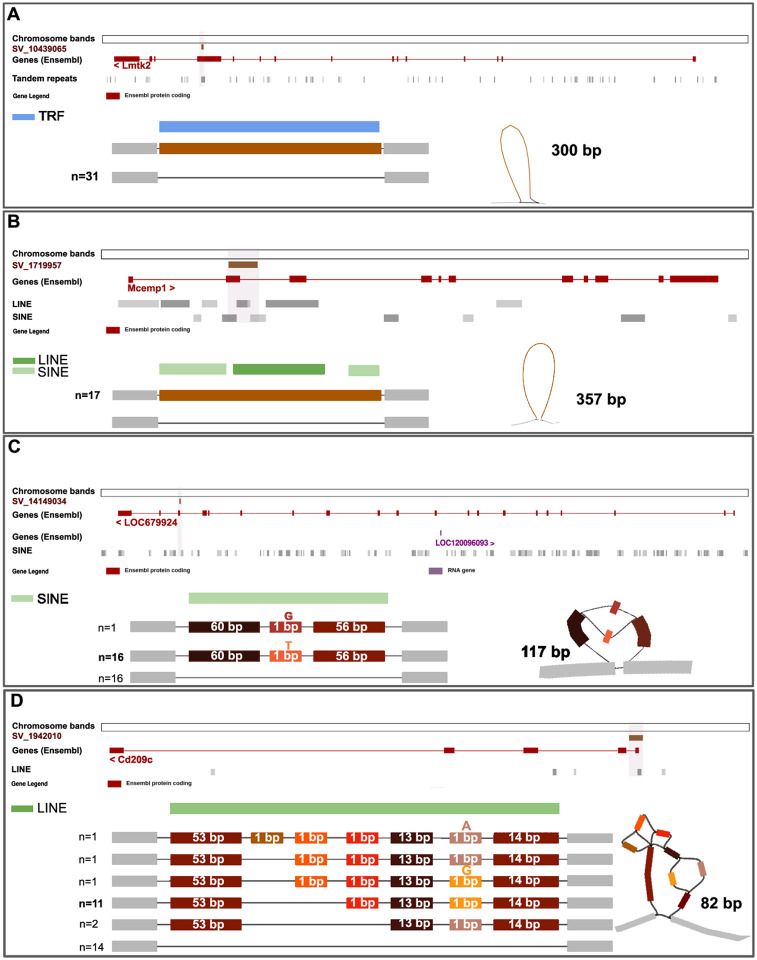
Figure 3. Resolution of complex haplotypes in validated structural variants (SVs) discovered using pangenome graphs.
Visualization using ODGI and Bandage of four SVs predicted to have high impact and validated with Oxford Nanopore sequencing technology. (A) A deletion of 300bp in the Lmtk2 gene supported by all four methods of identification, is found in all 31 samples and contains a Tandem Repeats element (TRF). (B) A deletion of 357bp in the Mcemp1 gene is supported by all four methods and is found in 17 rats. The gene contains SINE and LINE elements. (C) A deletion of 117 bp in the LOC679924 gene is supported by two methods, and includes a single nucleotide variant. The SV overlaps for 80 bp with a SINE element. (D) A complex insertion of 82 bp in the Cd209c gene, supported by all four methods. The insertion resolves in six haplotypes of which two are most abundant, and falls completely in a non-LTR retrotransposon (LINE).