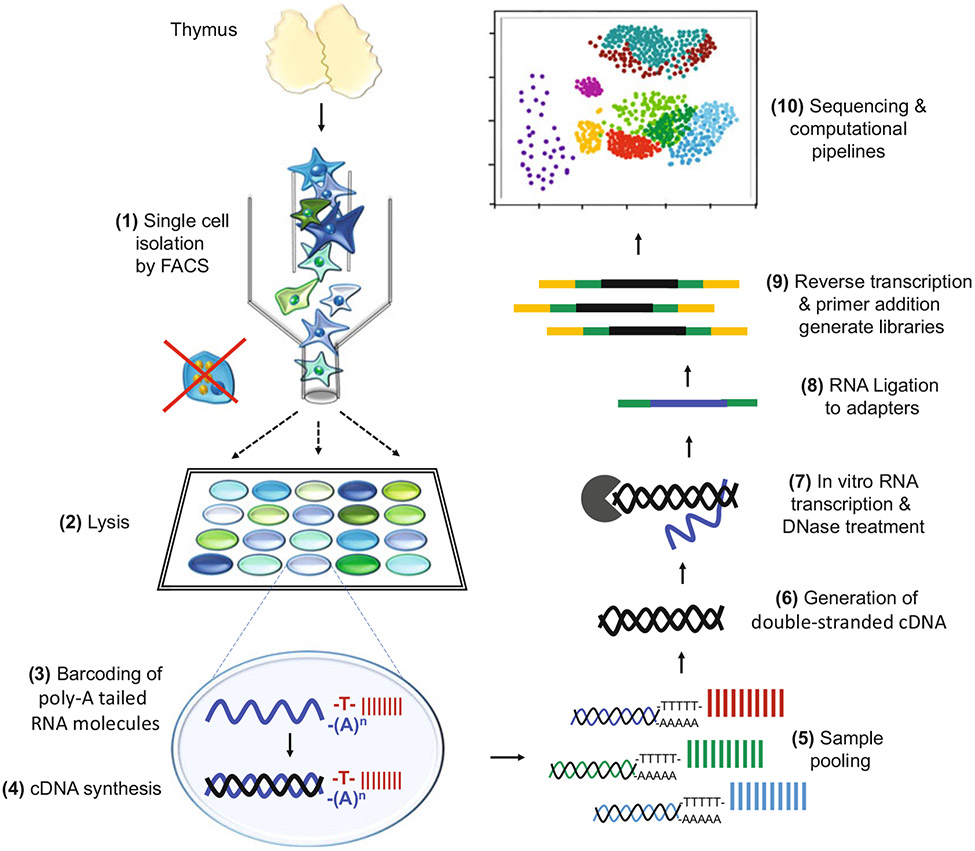
Fig. 2.
Schematic diagram of scRNA-Seq in the TEC scRNA-Seq framework based on the Massively parallel RNA single-cell sequencing framework (MARS-Seq) [93]. Thymic cells are used as an example. Single cells are first isolated by FACS eliminating cell complexes such as nurse cells. Next, cells are lysed in individual wells and, poly-A tailed RNA molecules are barcoded by annealing them to randomized molecular tags (unique molecular identifiers). Molecular tags also contain a T7 promoter (not shown), which will be used later for in vitro transcription. Then, reverse transcription is performed, generating the first cDNA strand. Next, individual cell lysates are pooled, and RNA-cDNA hybrids are converted to double-stranded cDNA (second strand synthesis). Then, in vitro transcription generates single-chain RNA molecules (this constitutes the first round of amplification), which are then fragmented and ligated to sequencing adapters. Finally, reverse transcription and a second round of amplification generate cDNA libraries for sequencing