Figure 3. Mechanism underlying YX968-induced degradation of HDAC3 and HDAC8.
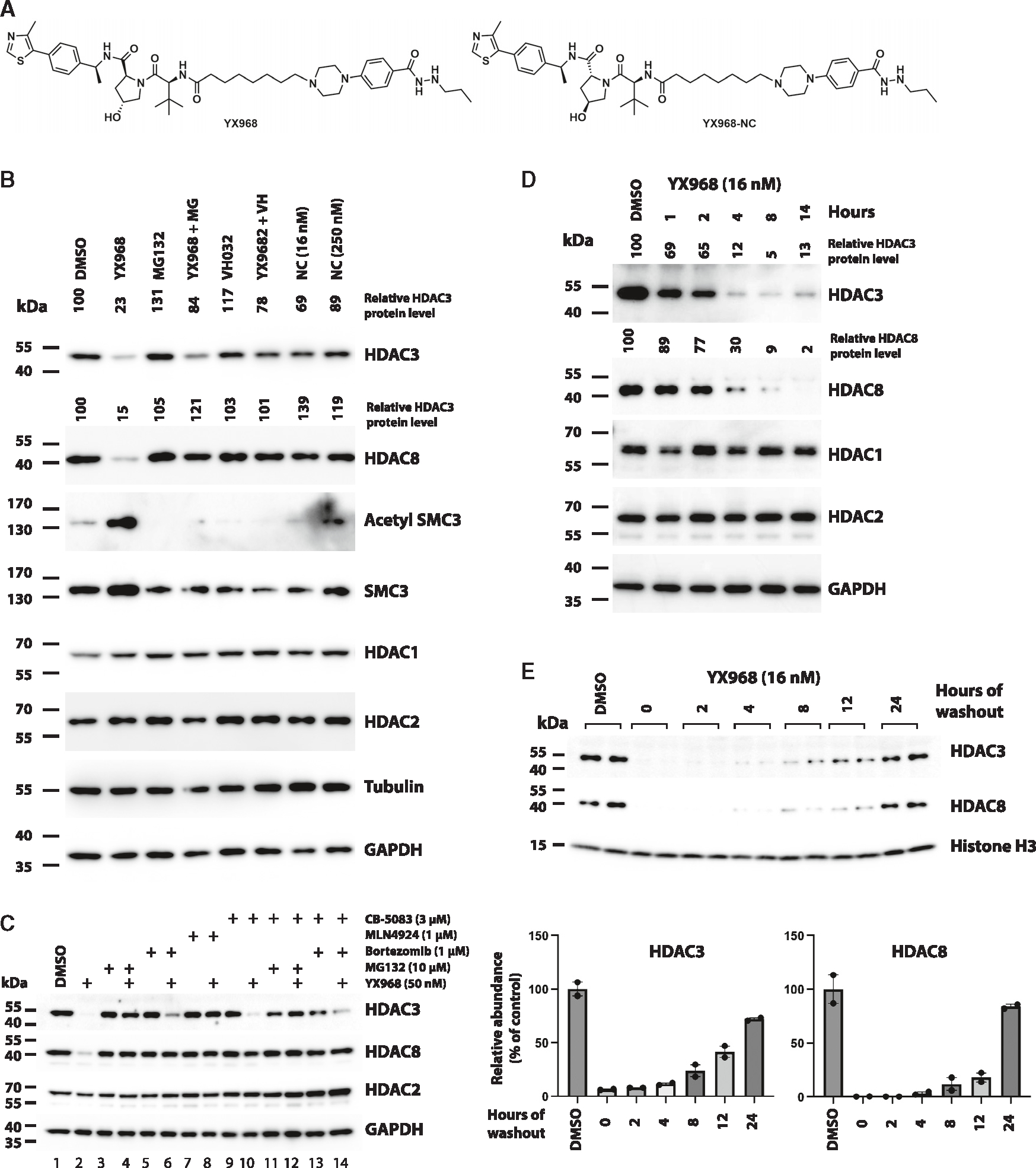
(A) Chemical structure of XY968 and YX968-NC (the negative control of YX968 that does not bind VHL).
(B) MDA-MB-231 cells were pretreated with 1 μM MG132 (MG), or 10 μM VHL ligand VH032 for 1 h followed by adding YX968 (16 nM) for 14 h. Cells were also similarly treated with YX968-NC (Shown as NC in Figure 3B). Western blot was done for detecting the indicated proteins.
(C) Additional probes for assessing the mechanism by which YX968 degrades HDAC3/8.
(D) Time-course assay in MDA-MB-231 cells after treatment with 16 nM YX968.
(E) MDA-MB-231 cells were incubated with 16 nM YX968 for 24 h followed by drug washout, and the treated cells were cultured in drug-free medium for the indicated time points. Samples from two independent assays are loaded. The normalized levels of HDAC3/8 protein are plotted. See also Figures S2 and S4.
