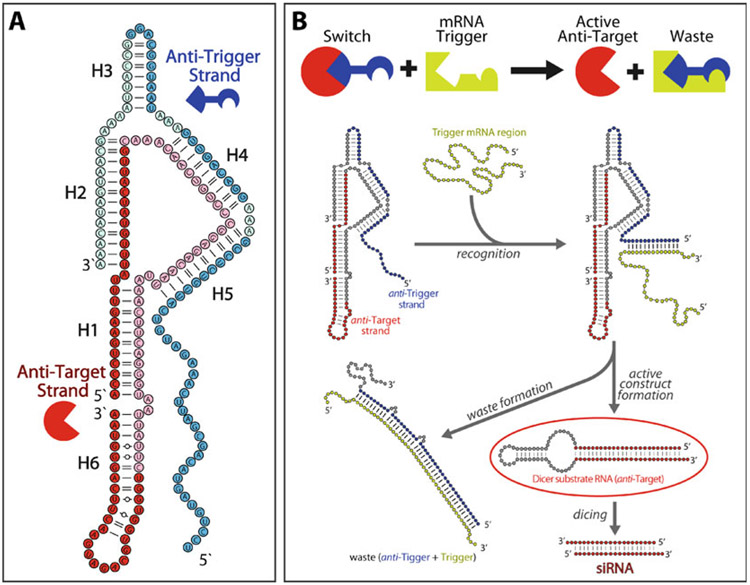
Fig. 1.
Design of a two-stranded RNA switch. (a) The two-stranded switch is assembled from an anti-trigger strand (blue) and an anti-target strand (red). The majority of the anti-trigger strand (dark blue nodes) is designed to be complementary to the trigger RNA sequence, while the 5′ and 3′ end regions of the anti-target strand (dark red nodes) encode siRNA sense and antisense sequences, respectively. (b) The RNA switch is initially in an inert conformation. Interaction of the anti-trigger’s single-stranded toehold with a trigger RNA induces a conformational change that releases the anti-target strand as an active shRNA-like Dicer substrate. Adapted and reprinted with permission from [18], copyright 2016 American Chemical Society