Abstract
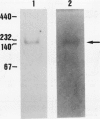
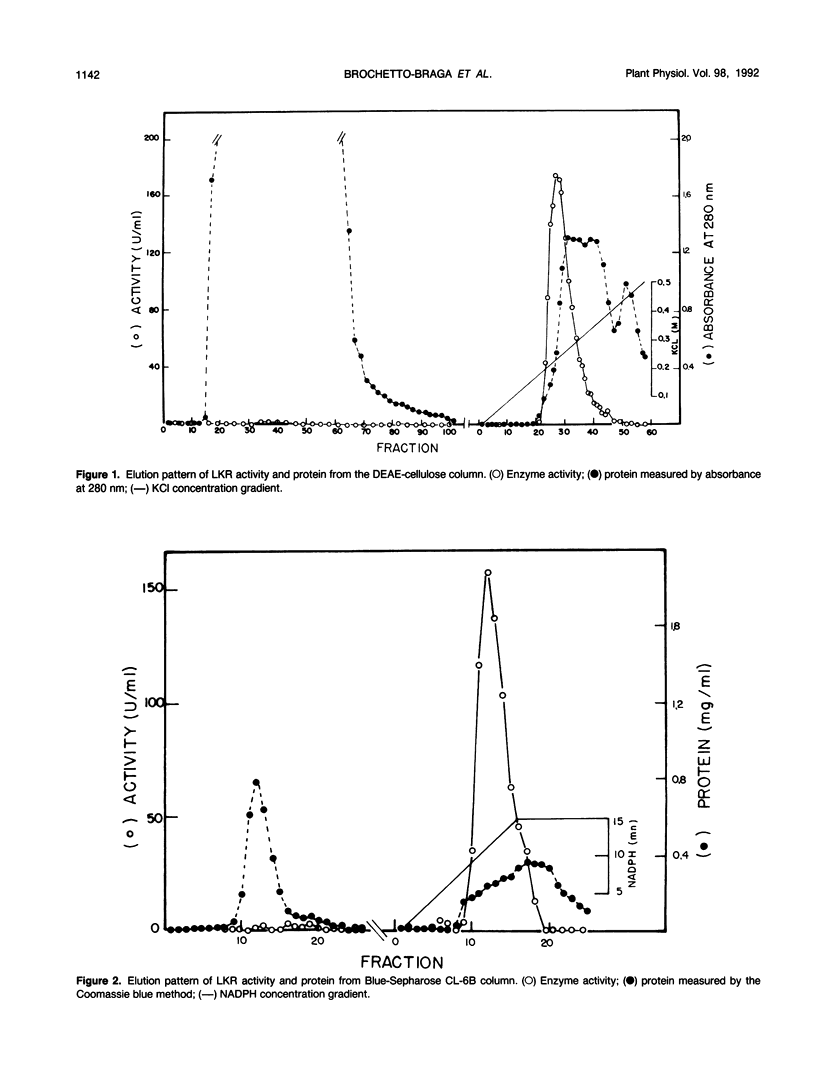
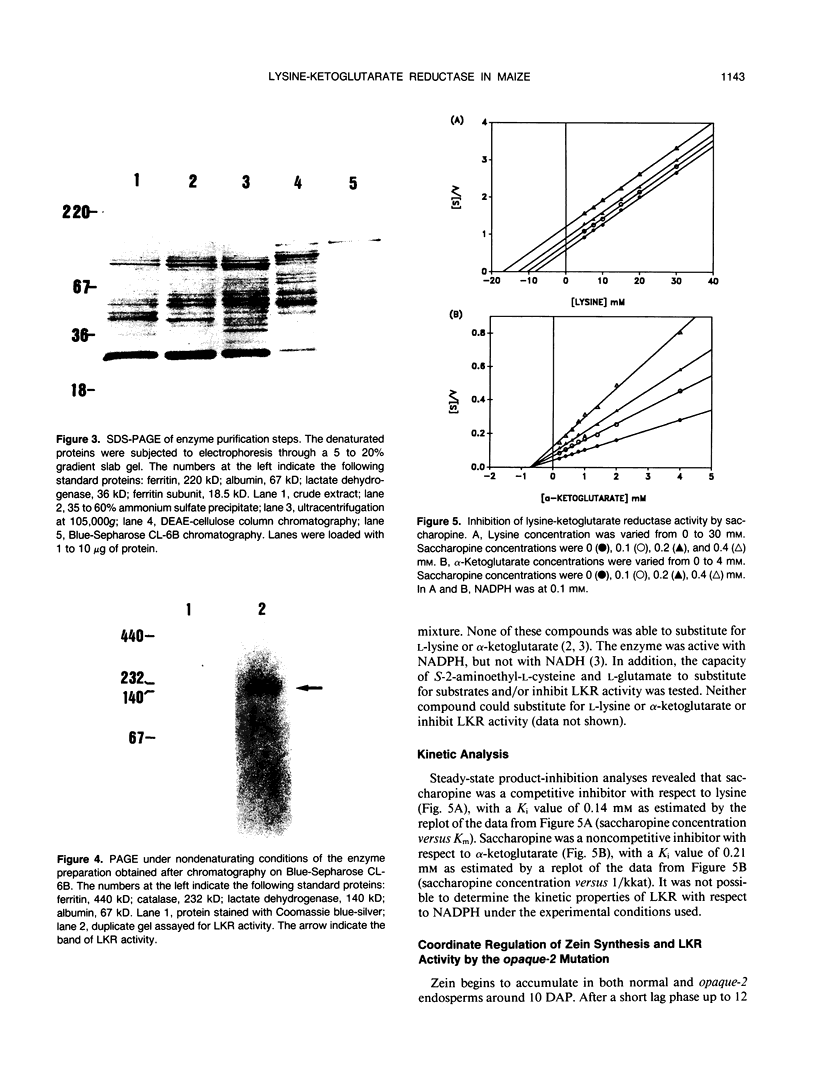
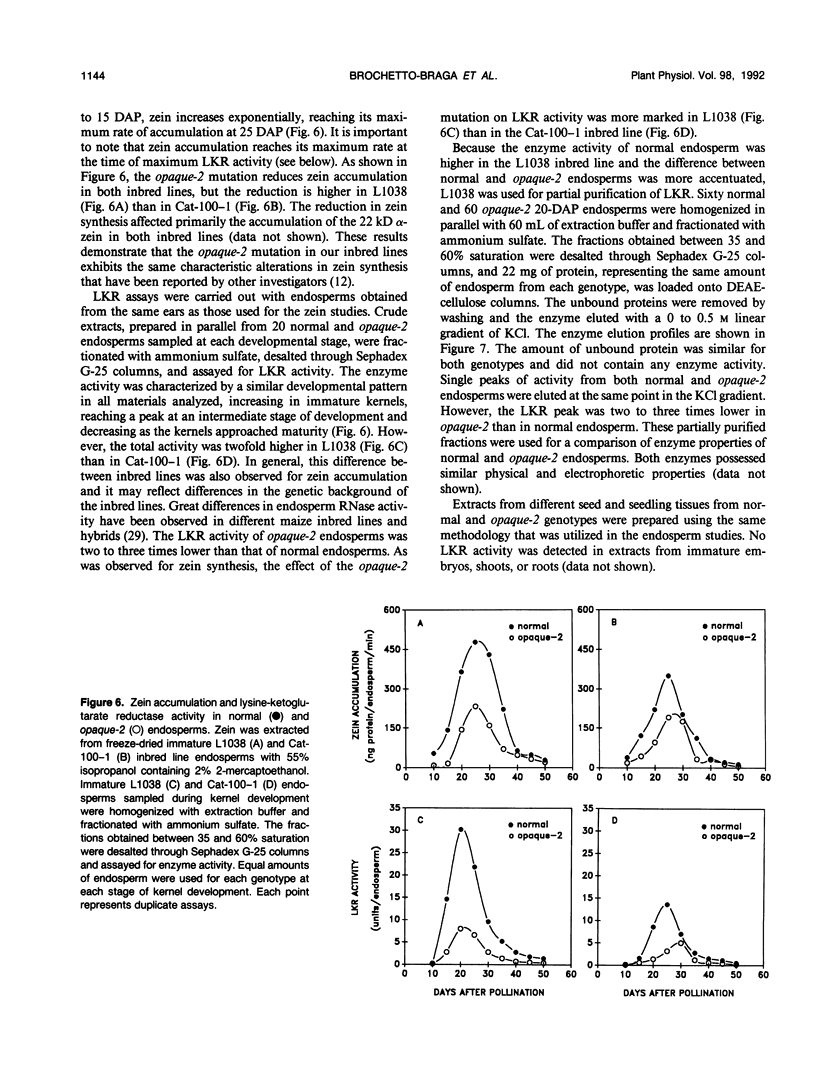
Lysine-ketoglutarate reductase catalyzes the first step of lysine catabolism in maize (Zea mays L.) endosperm. The enzyme condenses l-lysine and α-ketoglutarate into saccharopine using NADPH as cofactor. It is endosperm-specific and has a temporal pattern of activity, increasing with the onset of kernel development, reaching a peak 20 to 25 days after pollination, and there-after decreasing as the kernel approaches maturity. The enzyme was extracted from the developing maize endosperm and partially purified by ammonium-sulfate precipitation, anion-exchange chromatography on DEAE-cellulose, and affinity chromatography on Blue-Sepharose CL-6B. The preparation obtained from affinity chromatography was enriched 275-fold and had a specific activity of 411 nanomoles per minute per milligram protein. The native and denaturated enzyme is a 140 kilodalton protein as determined by polyacrylamide gel electrophoresis. The enzyme showed specificity for its substrates and was not inhibited by either aminoethyl-cysteine or glutamate. Steady-state product-inhibition studies revealed that saccharopine was a noncompetitive inhibitor with respect to α-ketoglutarate and a competitive inhibitor with respect to lysine. This is suggestive of a rapid equilibrium-ordered binding mechanism with a binding order of lysine, α-ketoglutarate, NADPH. The enzyme activity was investigated in two maize inbred lines with homozygous normal and opaque-2 endosperms. The pattern of lysine-ketoglutarate reductase activity is coordinated with the rate of zein accumulation during endosperm development. A coordinated regulation of enzyme activity and zein accumulation was observed in the opaque-2 endosperm as the activity and zein levels were two to three times lower than in the normal endosperm. Enzyme extracted from L1038 normal and opaque-2 20 days after pollination was partially purified by DEAE-cellulose chromatography. Both genotypes showed a similar elution pattern with a single activity peak eluted at approximately 0.2 molar KCL. The molecular weight and physical properties of the normal and opaque-2 enzymes were essentially the same. We suggest that the Opaque-2 gene, which is a transactivator of the 22 kilodalton zein genes, may be involved in the regulation of the lysine-ketoglutarate reductase gene in maize endosperm. In addition, the decreased reductase activity caused by the opaque-2 mutation may explain, at least in part, the elevated concentration of lysine found in the opaque-2 endosperm.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arruda P., Sodek L., da Silva W. J. Lysine-ketoglutarate reductase activity in developing maize endosperm. Plant Physiol. 1982 Apr;69(4):988–989. doi: 10.1104/pp.69.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A. B. In vivo incorporation of (14-C) lysine into the endosperm proteins of wild type and high-lysine barley. FEBS Lett. 1975 Apr 1;52(2):288–291. doi: 10.1016/0014-5793(75)80827-1. [DOI] [PubMed] [Google Scholar]
- De Moreno M. R., Smith J. F., Smith R. V. Silver staining of proteins in polyacrylamide gels: increased sensitivity through a combined Coomassie blue-silver stain procedure. Anal Biochem. 1985 Dec;151(2):466–470. doi: 10.1016/0003-2697(85)90206-4. [DOI] [PubMed] [Google Scholar]
- Dotson S. B., Somers D. A., Gengenbach B. G. Kinetic studies of lysine-sensitive aspartate kinase purified from maize suspension cultures. Plant Physiol. 1990 May;93(1):98–104. doi: 10.1104/pp.93.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjellstedt T. A., Robinson J. C. Purification and properties of L-lysine-alpha-ketoglutarate reductase from human placenta. Arch Biochem Biophys. 1975 Jun;168(2):536–548. doi: 10.1016/0003-9861(75)90285-4. [DOI] [PubMed] [Google Scholar]
- Frisch D. A., Gengenbach B. G., Tommey A. M., Sellner J. M., Somers D. A., Myers D. E. Isolation and characterization of dihydrodipicolinate synthase from maize. Plant Physiol. 1991 Jun;96(2):444–452. doi: 10.1104/pp.96.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings H., Maddaloni M., Lazzaroni N., Di Fonzo N., Motto M., Salamini F., Thompson R. The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 1989 Oct;8(10):2795–2801. doi: 10.1002/j.1460-2075.1989.tb08425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodrzycki R., Boston R. S., Larkins B. A. The opaque-2 mutation of maize differentially reduces zein gene transcription. Plant Cell. 1989 Jan;1(1):105–114. doi: 10.1105/tpc.1.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmer S., Maddaloni M., Motto M., Di Fonzo N., Hartings H., Salamini F., Thompson R. D. The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J. 1991 Mar;10(3):617–624. doi: 10.1002/j.1460-2075.1991.tb07989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MERTZ E. T., BATES L. S., NELSON O. E. MUTANT GENE THAT CHANGES PROTEIN COMPOSITION AND INCREASES LYSINE CONTENT OF MAIZE ENDOSPERM. Science. 1964 Jul 17;145(3629):279–280. doi: 10.1126/science.145.3629.279. [DOI] [PubMed] [Google Scholar]
- Møller B. L. Lysine Catabolism in Barley (Hordeum vulgare L.). Plant Physiol. 1976 May;57(5):687–692. doi: 10.1104/pp.57.5.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIGAM S. N., McCONNELL W. B. Studies on wheat plants using carbon-14 compounds. XIX. Observations on the metabolism of lysine-C14. Can J Biochem Physiol. 1963 Jun;41:1367–1371. [PubMed] [Google Scholar]
- Schmidt R. J., Burr F. A., Aukerman M. J., Burr B. Maize regulatory gene opaque-2 encodes a protein with a "leucine-zipper" motif that binds to zein DNA. Proc Natl Acad Sci U S A. 1990 Jan;87(1):46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. J., Burr F. A., Burr B. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science. 1987 Nov 13;238(4829):960–963. doi: 10.1126/science.2823388. [DOI] [PubMed] [Google Scholar]
- Soave C., Tardani L., Di Fonzo N., Salamini F. Zein level in maize endosperm depends on a protein under control of the opaque-2 and opaque-6 loci. Cell. 1981 Dec;27(2 Pt 1):403–410. doi: 10.1016/0092-8674(81)90423-2. [DOI] [PubMed] [Google Scholar]
- Sodek L., Wilson C. M. Incorporation of leucine-14C and lysine-14C into protein in the developing endosperm of normal and opaque-2 corn. Arch Biochem Biophys. 1970 Sep;140(1):29–38. doi: 10.1016/0003-9861(70)90006-8. [DOI] [PubMed] [Google Scholar]
- Spector T. Refinement of the coomassie blue method of protein quantitation. A simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 microgram of protein. Anal Biochem. 1978 May;86(1):142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- Susor W. A., Rutter W. J. Method for the detection of pyruvate kinase, aldolase, and other pyridine nucleotide linked enzyme activities after electrophoresis. Anal Biochem. 1971 Sep;43(1):147–155. doi: 10.1016/0003-2697(71)90119-9. [DOI] [PubMed] [Google Scholar]
- Wilson C. M., Alexander D. E. Ribonuclease activity in normal and opaque-2 mutant endosperm of maize. Science. 1967 Mar 24;155(3769):1575–1576. doi: 10.1126/science.155.3769.1575. [DOI] [PubMed] [Google Scholar]
- Wilson C. M. Plant Nucleases: VI. GENETIC AND DEVELOPMENTAL VARIABILITY IN RIBONUCLEASE ACTIVITY IN INBRED AND HYBRID CORN ENDOSPERMS. Plant Physiol. 1980 Jul;66(1):119–125. doi: 10.1104/pp.66.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]