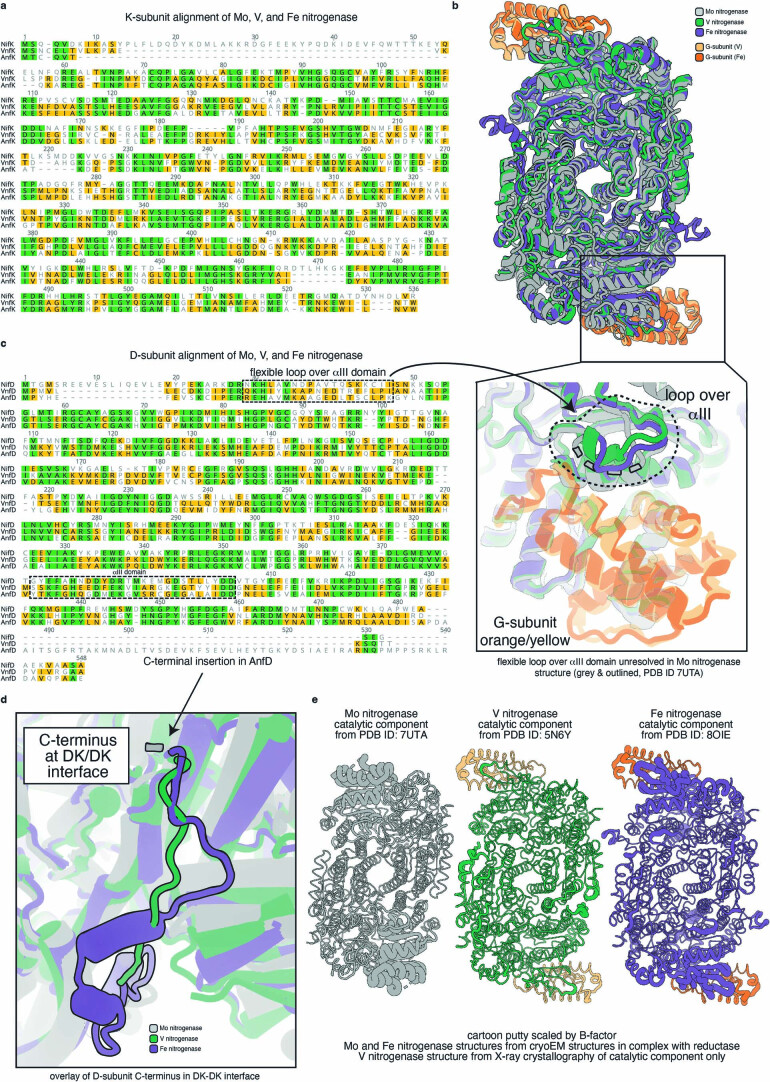
Extended Data Fig. 4. Sequence and structural alignment of Mo, V, and Fe nitrogenase catalytic components.
(a) Sequence alignment of Azotobacter vinelandii Mo and V nitrogenase K-subunit (NifK and VnfK) to Rhodobacter capsulatus Fe nitrogenase K-subunit (AnfK). Identical sites are shown in green and similar sites (BLOSUM62 matrix68 threshold of 2) that occur in 2 of 3 sequences are highlighted in yellow. (b) Structural alignment of the catalytic components of the Mo (PDB: 7UTA, resting-state catalytic component), V (PDB: 5N6Y, resting-state catalytic component), and Fe (PDB: 8OIE, AlF3-trapped complex) nitrogenases aligned in (a) and including the G-subunit for V and Fe nitrogenase. Inset at bottom shows a zoomed view of the αIII domain, with arrows depicting the origin in the sequence alignment. (c) Sequence alignment of Azotobacter vinelandii Mo and V nitrogenase D-subunit (NifD and VnfD) to Rhodobacter capsulatus Fe nitrogenase D-subunit (AnfD). Identical sites are shown in green and similar sites (BLOSUM62 matrix, threshold of 2) that occur in 2 of 3 sequences are highlighted in yellow. (d) Close-up view into the interface between individual DK-halves of the catalytic component. Arrow points towards C-terminus of D-subunit, which is extended in Fe nitrogenase. (e) Structural alignment of putty-styled cartoon Mo, V, and Fe nitrogenases (same models as in (b)). Putty size is scaled by B-factor. Mo (PDB: 7UTA, resting-state catalytic component) and Fe (PDB: 8OIE, AlF3-trapped complex) nitrogenase structures are measured by cryo-EM, whereas V (PDB: 5N6Y, resting-state catalytic component) nitrogenase structure derives from X-ray crystallography.