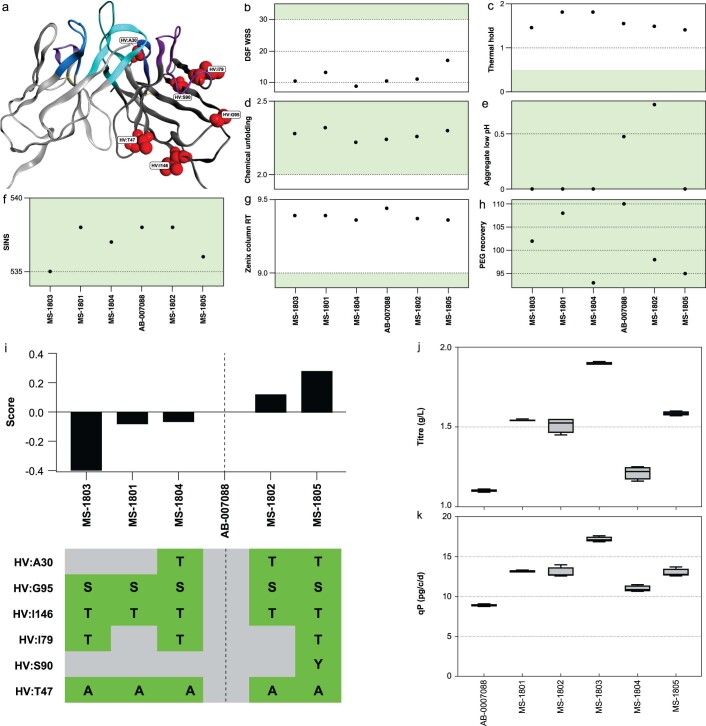
Extended Data Fig. 5. Generation and developability properties of engineered variants from backup molecule.
a, Mutations made to generate engineered variants are depicted in the Fv region of a Fab structure model of AB-007088 built in the MOE Antibody Modeler (see Methods). The light chain framework is in silver and the heavy chain in dim grey. The CDRs for light and heavy chains are indicated, respectively, as CDR1 (light and dark blue), CDR2 (light and dark purple), and CR3 (light and dark cyan). Mutation sites of engineered variants are labelled as stability violations (red, see Methods). b–h, Developability properties of engineered variants of parental backup, AB-007088, as characterized by scores from, b, DSF WSS, c, thermal hold, d, chemically induced unfolding, e, low pH stability (aggregate low pH hold), f, self-interaction nanoparticle spectroscopy (SINS), g, stand-up monolayer affinity chromatography (Zenix column RT), h, relative solubility analysis (PEG recovery) with green shading that indicates preferred ranges. i, Aggregate score of assay panel results from b-h for each engineered variant with mutations shown in comparison to AB-007088. j–k, Characterization of stable cell production pools generated from engineered variants compared to AB-007088 for, j, production titres, (Titre, g/L) and, k, cell-specific productivity (qP, pg per cell per day). Boxes indicate interquartile ranges, lines within boxes are medians, and whiskers represent minimum and maximum (4 independent replicates of each antibody).