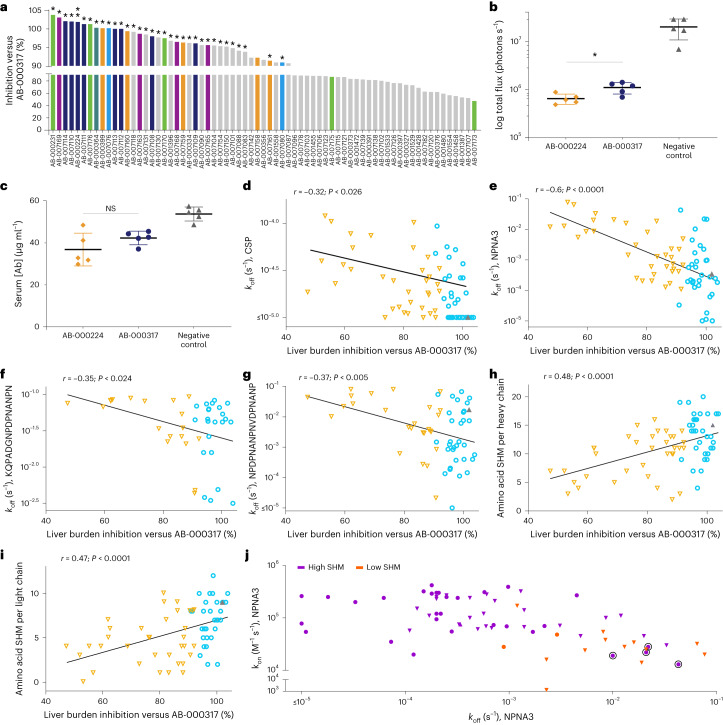
Fig. 3. In vitro binding, in vivo activity and SHM associated with developable mAbs.
a–c, Liver burden model. a, Percentage inhibition compared with untreated, infected mice (geometric mean, n = 5, 100 μg per mouse) normalized to the activity of AB-000317 of n = 69 antibodies (32 lineages, *P > 0.05, **P < 0.05 activity greater than AB-000317, no icon P < 0.05 activity less than AB-000317, two-tailed, nonparametric log-rank), with colors other than gray indicating the six lineages containing the most efficacious mAbs. b,c, Example data from AB-000224 and AB-000317 of parasite bioluminescence in the liver (total flux, photons s−1), *P = 0.03 (b) and serum concentrations (serum(Ab), μg ml−1) of mAb at the time of sporozoite challenge (c), P > 0.2 (NS), mean ± s.d. (n = 5 mice), two-tailed Mann–Whitney test. d–i, Simple two-tailed linear regression of percentage liver burden inhibitory activity compared with untreated, infected mice (geometric mean, n = 5) and normalized to the activity of AB-000317, with each mAb indicated as having activity either significantly better (gray upward-pointing triangle), not different to (cyan circles) or weaker than (orange downward-pointing triangles) AB-000317 (two-tailed, nonparametric log-rank) versus d–g, log-transformed binding off-rate (koff) against CSP (n = 70) (d), major repeat (NPNA3, n = 70) (e), junctional (KQPADGNPDPNANPN, n = 42) (f) and short minor-repeat (NPDPNANPNVDPNANP, n = 60) peptides (g) and versus h,i, the number of amino acid changes from germline (SHM) for each mAb (n = 69)—heavy (h) and light chain (i) (see Extended Data Table 1 for correlation analyses of non-log-transformed data). Dissociation rate (koff) measurements were limited to a minimum of 10−5 s−1 and mAbs were excluded from correlation analyses if assigned this value. j, log-transformed SPR binding off- (koff) versus on-rates (kon) against NPNA3 peptide of mAbs with either high SHM (purple, ≥20 nucleotide mutations per clone, n = 55) or low SHM (orange, <20 nucleotide mutations per clone, n = 14) and with activity weaker (downward-pointing triangles), not different to (circles) or better than (upward-pointing triangle) AB-000317 (two-tailed, nonparametric log-rank). mAbs from a lineage reported to bind CSP with Fab–Fab homotypic interactions are indicated (black circles, AB-000399 (refs. 41,42), AB-007159, AB-007160, AB-007161).