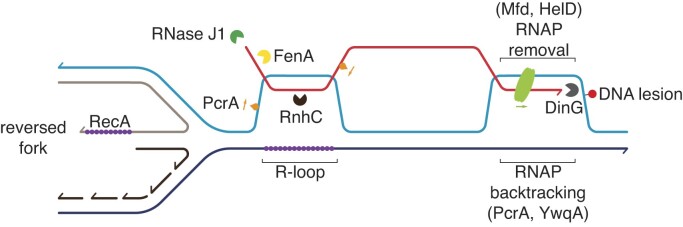
Figure 4.
Cartoon showing how B. subtilis proteins may contribute to resolve RTCs. Here, a CD RTC is illustrated. A replisome clashes with multiple RNAPs transcribing highly expressed genes (i.e. rRNA genes), or the RNAP finds a DNA lesion on the template strand (red circle), and transcription is halted. Upon that, the stalled fork has a tendency to reverse and RNAP to backtrack, and this causes topological constrains that facilitate R-loop formation. RecA (purple circles) may bind to the ssDNA region in the regressed fork or in the R-loop. RNase J1 (green Pac-Man) or FenA (yellow Pac-Man) degrades the mRNA. RNAP and RecA, acting as hubs, interact with and recruit PcrA (orange drop), RnhC (black Pac-Man), or DinG (grey Pac-Man) at the trafficking conflict. PcrA displaces and RnhC degrades the RNA strand of the R-loop. PcrA or YwqA facilitates RNAP backtracking, Mfd or HelD facilitates RNAP removal and DinG degrades the exposed 3′-end of the mRNA to facilitate transcription reinitiation upon removal of the lesions. RecA bound to the reversed fork may protect it from degradation. Although not depicted here, topoisomerases may also play a role at RTC sites.