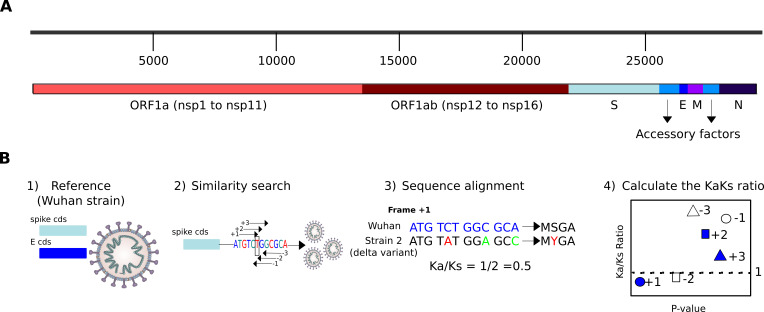
Fig 1.
Viral genes and protocol for calculating the selection pressure. (A) SARS-CoV-2 genome organization. The genome is divided into non-structural genes (nsp genes coming from both ORF1a and ORF1ab), structural genes (S, E, M, and N), and accessory factors. (B) Procedure for calculating the Ka/Ks ratio: (1) CDSs are obtained from the reference strain (GenBank: MN908947.3). (2) The six putative reading frames for each CDS in the reference strain are extracted, and homologous sequences are searched for in all the strains. (3) The Ka/Ks ratio for each frame can then be calculated using pairwise alignments of the homologs (the same genes from the other viral strains). The analyzed gene is highlighted in blue, and nucleotide changes or in red (when they correspond to nonsynonymous changes), or in green (to synonymous changes). (4) Finally, the distribution of Ka/Ks ratios from all the viral genes can be shown, where we expect a value <1 for most of the genes in the frame +1 (negative selection), and slightly higher values in the frame −2. However, the other four frames should show values >1 (positive selection).