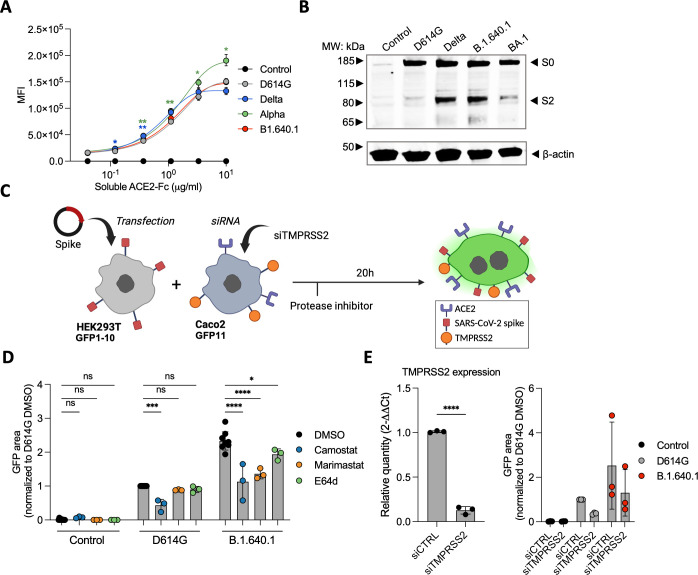
Fig 7.
The impact of host cell proteases on B.1.640.1 Spike fusion. (A) Staining of HEK293T cells transfected with Spike or control plasmids and stained with a serial dilution of soluble human ACE2-Fc followed by staining with anti-human-Fc secondary antibody. A two-way ANOVA test was performed with Geisser-Greenhouse correction to compare D614G to the respective variants, *P < 0.05, **P < 0.001. (B) S1/S2 cleavage of respective variant Spike proteins measured by western blot. HEK293T cells transfected with respective Spikes or control plasmids and cell lysates were analyzed 24 hours later (n = 2). (C) Schematic of HEK293T-GFP1-10 cells transfected with D614G Spike, B.1.640.1 Spike, or control plasmids and co-cultured with Caco-2-GFP11 in the presence of protease inhibitors. (D) Fusion of D614G and B.1.640.1 Spikes, quantified by GFP area, in the presence 10 mM camostat, marimastat, E64d, or a DMSO control. (E) (Left) Total RNA was extracted from Caco2 cells treated with TMPRSS2 siRNA or siCTRL, and RT-qPCR was performed. Data were normalized to β-Tubulin levels. Relative mRNA expression normalized to siCtrl condition (2−ΔΔCT) was plotted. (Right) Fusion of D614G and B.1.640.1, quantified by GFP, in the presence of TMPRSS2 knockdown or wild-type Caco-2 cells. For (D) and (E), ordinary one-way ANOVA tests were performed with Tukey’s multiple comparison test to compare D614G to respective variants, *P < 0.05, ***P < 0.0001, ****P < 0.00001. Error bars represent SD.